Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
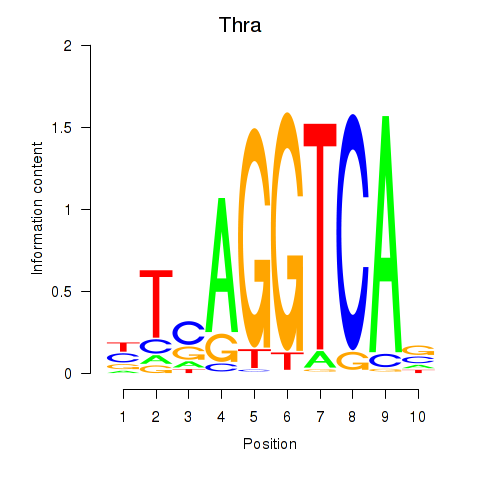
Results for Thra
Z-value: 0.77
Transcription factors associated with Thra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Thra
|
ENSRNOG00000009066 | thyroid hormone receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Thra | rn6_v1_chr10_+_86657285_86657285 | 0.19 | 5.1e-04 | Click! |
Activity profile of Thra motif
Sorted Z-values of Thra motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_33606124 | 30.28 |
ENSRNOT00000065210
|
AC115371.1
|
|
| chr1_+_80321585 | 29.80 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr11_+_80736576 | 27.48 |
ENSRNOT00000047678
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr15_+_33632416 | 27.39 |
ENSRNOT00000068212
|
AC130940.1
|
|
| chr8_+_50559126 | 27.28 |
ENSRNOT00000024918
|
Apoa5
|
apolipoprotein A5 |
| chr12_+_45905371 | 21.29 |
ENSRNOT00000039275
|
Hspb8
|
heat shock protein family B (small) member 8 |
| chr1_+_215609036 | 19.20 |
ENSRNOT00000076187
|
Tnni2
|
troponin I2, fast skeletal type |
| chr7_-_80796670 | 18.43 |
ENSRNOT00000010539
|
Abra
|
actin-binding Rho activating protein |
| chr10_+_82775691 | 16.32 |
ENSRNOT00000030737
|
Hils1
|
histone linker H1 domain, spermatid-specific 1 |
| chr4_-_145948996 | 16.03 |
ENSRNOT00000043476
|
Atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr10_-_18443934 | 15.04 |
ENSRNOT00000059895
ENSRNOT00000080021 |
Ranbp17
|
RAN binding protein 17 |
| chr17_+_76002275 | 14.78 |
ENSRNOT00000092665
ENSRNOT00000086701 |
Echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr9_+_92435896 | 13.13 |
ENSRNOT00000022901
|
Fbxo36
|
F-box protein 36 |
| chr1_+_72425707 | 12.63 |
ENSRNOT00000058956
|
Sbk2
|
SH3 domain binding kinase family, member 2 |
| chr5_-_76039760 | 12.20 |
ENSRNOT00000019959
|
RGD1306148
|
similar to KIAA0368 |
| chr2_+_229196616 | 11.78 |
ENSRNOT00000012773
|
Ndst4
|
N-deacetylase and N-sulfotransferase 4 |
| chr1_-_89488223 | 11.63 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr8_+_50537009 | 11.62 |
ENSRNOT00000080658
|
Apoa4
|
apolipoprotein A4 |
| chr1_+_177048655 | 11.01 |
ENSRNOT00000081595
|
Mical2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr12_+_24978483 | 10.88 |
ENSRNOT00000040069
|
Eln
|
elastin |
| chr11_-_83905889 | 10.33 |
ENSRNOT00000075606
|
Fam131a
|
family with sequence similarity 131, member A |
| chr11_-_82884660 | 10.31 |
ENSRNOT00000073526
|
LOC100912498
|
protein FAM131A-like |
| chr1_+_137799185 | 10.03 |
ENSRNOT00000083590
ENSRNOT00000092778 |
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr7_-_123586919 | 9.93 |
ENSRNOT00000011484
|
Ndufa6
|
NADH:ubiquinone oxidoreductase subunit A6 |
| chr17_+_77167740 | 9.85 |
ENSRNOT00000042881
|
Optn
|
optineurin |
| chr11_-_4332255 | 9.64 |
ENSRNOT00000087133
|
Cadm2
|
cell adhesion molecule 2 |
| chr1_-_66212418 | 9.61 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr16_+_23553647 | 9.28 |
ENSRNOT00000041994
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr17_+_36334147 | 9.27 |
ENSRNOT00000050261
|
E2f3
|
E2F transcription factor 3 |
| chr11_+_64601029 | 9.15 |
ENSRNOT00000004138
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr3_-_2727616 | 9.12 |
ENSRNOT00000061904
|
C8g
|
complement C8 gamma chain |
| chr6_+_2216623 | 8.74 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr20_+_5008508 | 8.69 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr13_+_51034256 | 8.68 |
ENSRNOT00000004528
ENSRNOT00000046854 ENSRNOT00000087320 |
Mybph
|
myosin binding protein H |
| chr10_+_39435227 | 8.21 |
ENSRNOT00000042144
ENSRNOT00000077185 |
P4ha2
|
prolyl 4-hydroxylase subunit alpha 2 |
| chr15_-_3544685 | 7.69 |
ENSRNOT00000015179
ENSRNOT00000085126 |
Vcl
|
vinculin |
| chr1_+_84067938 | 7.57 |
ENSRNOT00000057213
|
Numbl
|
NUMB-like, endocytic adaptor protein |
| chr1_+_228142778 | 7.53 |
ENSRNOT00000028517
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr2_+_98252925 | 7.48 |
ENSRNOT00000011579
|
Pex2
|
peroxisomal biogenesis factor 2 |
| chr6_-_122721496 | 7.45 |
ENSRNOT00000079697
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr7_-_33916338 | 7.27 |
ENSRNOT00000005492
|
Cfap54
|
cilia and flagella associated protein 54 |
| chr6_-_135829953 | 7.06 |
ENSRNOT00000080623
ENSRNOT00000039059 |
Cdc42bpb
|
CDC42 binding protein kinase beta |
| chr3_+_37545238 | 6.43 |
ENSRNOT00000070792
|
Tnfaip6
|
TNF alpha induced protein 6 |
| chr3_-_134406576 | 6.10 |
ENSRNOT00000081589
|
Sel1l2
|
SEL1L2 ERAD E3 ligase adaptor subunit |
| chr3_+_148510779 | 6.03 |
ENSRNOT00000012156
|
Xkr7
|
XK related 7 |
| chrX_+_68752597 | 5.54 |
ENSRNOT00000077039
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr13_-_74740458 | 5.24 |
ENSRNOT00000006548
|
Tex35
|
testis expressed 35 |
| chr4_-_99746560 | 5.18 |
ENSRNOT00000012021
|
Mrpl35
|
mitochondrial ribosomal protein L35 |
| chr1_+_250426158 | 5.08 |
ENSRNOT00000067643
|
A1cf
|
APOBEC1 complementation factor |
| chr3_+_143129248 | 4.95 |
ENSRNOT00000006667
|
Cst8
|
cystatin 8 |
| chr14_+_12218553 | 4.81 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr18_+_30574627 | 4.72 |
ENSRNOT00000060484
|
Pcdhb19
|
protocadherin beta 19 |
| chr1_-_59732409 | 4.67 |
ENSRNOT00000014824
|
Has1
|
hyaluronan synthase 1 |
| chr1_-_80716143 | 4.45 |
ENSRNOT00000092048
ENSRNOT00000025829 |
Cblc
|
Cbl proto-oncogene C |
| chr6_+_7961413 | 4.35 |
ENSRNOT00000007638
ENSRNOT00000061871 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr9_+_111597037 | 4.34 |
ENSRNOT00000021758
|
Fer
|
FER tyrosine kinase |
| chr8_-_47339343 | 4.31 |
ENSRNOT00000081007
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr4_-_45332420 | 4.27 |
ENSRNOT00000083039
|
Wnt2
|
wingless-type MMTV integration site family member 2 |
| chr3_+_95715193 | 4.13 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr6_+_112203679 | 4.13 |
ENSRNOT00000031205
|
Nrxn3
|
neurexin 3 |
| chr19_+_37675113 | 4.11 |
ENSRNOT00000024125
|
RGD1561415
|
RGD1561415 |
| chr20_+_31102476 | 4.03 |
ENSRNOT00000078719
|
Lrrc20
|
leucine rich repeat containing 20 |
| chrX_+_54734385 | 4.02 |
ENSRNOT00000005023
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr1_-_169334093 | 3.98 |
ENSRNOT00000032587
|
Ubqln3
|
ubiquilin 3 |
| chr3_-_60813869 | 3.96 |
ENSRNOT00000058234
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr5_+_14415606 | 3.95 |
ENSRNOT00000089273
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr10_+_85608544 | 3.89 |
ENSRNOT00000092122
|
Mllt6
|
MLLT6, PHD finger domain containing |
| chr1_+_82174451 | 3.86 |
ENSRNOT00000027783
|
Tmem145
|
transmembrane protein 145 |
| chr5_+_58416432 | 3.65 |
ENSRNOT00000039366
|
LOC100360821
|
rCG55159-like |
| chr10_+_72486152 | 3.56 |
ENSRNOT00000004193
|
RGD1310166
|
similar to Chromodomain-helicase-DNA-binding protein 1 (CHD-1) |
| chr20_+_27352276 | 3.40 |
ENSRNOT00000076534
|
Rn50_20_0292.1
|
|
| chr5_+_173274774 | 3.28 |
ENSRNOT00000025952
|
Ccnl2
|
cyclin L2 |
| chr10_-_85435016 | 3.27 |
ENSRNOT00000079921
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr1_-_214414763 | 3.24 |
ENSRNOT00000025240
ENSRNOT00000077776 |
LOC100911440
|
mitochondrial glutamate carrier 1-like |
| chr9_+_93445002 | 3.17 |
ENSRNOT00000025029
|
LOC501180
|
similar to hypothetical protein MGC35154 |
| chrX_-_107442878 | 3.09 |
ENSRNOT00000052302
|
Glra4
|
glycine receptor, alpha 4 |
| chr6_+_137243185 | 3.06 |
ENSRNOT00000030879
|
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr7_-_122926336 | 3.05 |
ENSRNOT00000000205
|
Chadl
|
chondroadherin-like |
| chr1_+_268189277 | 3.01 |
ENSRNOT00000065001
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr9_+_88494676 | 2.98 |
ENSRNOT00000089451
|
LOC102556337
|
mitochondrial fission factor-like |
| chr8_+_116332796 | 2.92 |
ENSRNOT00000021408
|
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr7_-_20070772 | 2.91 |
ENSRNOT00000071008
|
LOC102551633
|
sperm motility kinase W-like |
| chr3_-_2845593 | 2.85 |
ENSRNOT00000067479
|
Tmem141
|
transmembrane protein 141 |
| chr10_+_85257876 | 2.78 |
ENSRNOT00000014752
|
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr6_-_114488880 | 2.71 |
ENSRNOT00000087560
|
AC118957.1
|
|
| chr5_+_14415841 | 2.63 |
ENSRNOT00000010682
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr4_+_57715946 | 2.35 |
ENSRNOT00000079059
ENSRNOT00000034429 |
Klhdc10
|
kelch domain containing 10 |
| chr3_-_2434257 | 2.23 |
ENSRNOT00000013278
|
Stpg3
|
sperm-tail PG-rich repeat containing 3 |
| chr3_+_140024043 | 2.14 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
| chr10_-_87521514 | 2.13 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr5_+_147692391 | 2.04 |
ENSRNOT00000064855
|
Fam229a
|
family with sequence similarity 229, member A |
| chr10_-_102423084 | 2.01 |
ENSRNOT00000065145
|
Sdk2
|
sidekick cell adhesion molecule 2 |
| chr19_-_42180981 | 1.95 |
ENSRNOT00000019577
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr4_-_71958774 | 1.93 |
ENSRNOT00000024076
|
Olr804
|
olfactory receptor 804 |
| chr15_-_86142672 | 1.85 |
ENSRNOT00000057830
|
Commd6
|
COMM domain containing 6 |
| chr10_-_65329279 | 1.64 |
ENSRNOT00000088721
ENSRNOT00000078757 ENSRNOT00000014104 ENSRNOT00000014118 |
Flot2
|
flotillin 2 |
| chr4_-_145487426 | 1.56 |
ENSRNOT00000013488
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr10_-_87535438 | 1.51 |
ENSRNOT00000086873
|
Krtap2-4
|
keratin associated protein 2-4 |
| chr10_-_91807937 | 1.45 |
ENSRNOT00000005074
|
Wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr15_-_34444244 | 1.40 |
ENSRNOT00000027612
|
Cideb
|
cell death-inducing DFFA-like effector b |
| chr6_-_115616766 | 1.39 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr12_+_49578633 | 1.37 |
ENSRNOT00000072683
|
Crybb2
|
crystallin, beta B2 |
| chr1_-_168144280 | 1.36 |
ENSRNOT00000020969
|
Olr69
|
olfactory receptor 69 |
| chr20_+_5815837 | 1.32 |
ENSRNOT00000036999
|
Lhfpl5
|
lipoma HMGIC fusion partner-like 5 |
| chr6_+_136279496 | 1.24 |
ENSRNOT00000091946
|
Apopt1
|
apoptogenic 1, mitochondrial |
| chr10_-_12386222 | 1.21 |
ENSRNOT00000060994
|
Olr1364
|
olfactory receptor 1364 |
| chr16_-_69242028 | 1.19 |
ENSRNOT00000019002
|
Zfp703
|
zinc finger protein 703 |
| chr1_+_72635267 | 1.03 |
ENSRNOT00000090636
|
Il11
|
interleukin 11 |
| chr2_-_30127269 | 0.97 |
ENSRNOT00000023869
|
Cartpt
|
CART prepropeptide |
| chr15_+_86153628 | 0.78 |
ENSRNOT00000012842
|
Uchl3
|
ubiquitin C-terminal hydrolase L3 |
| chr1_-_64446818 | 0.75 |
ENSRNOT00000081980
|
Myadm
|
myeloid-associated differentiation marker |
| chr6_+_110624856 | 0.69 |
ENSRNOT00000014017
|
Vash1
|
vasohibin 1 |
| chr19_-_27535792 | 0.67 |
ENSRNOT00000024040
|
Olr1666
|
olfactory receptor 1666 |
| chr7_+_139444994 | 0.66 |
ENSRNOT00000090079
|
Tmem106c
|
transmembrane protein 106C |
| chr4_+_35279063 | 0.60 |
ENSRNOT00000011833
|
Nxph1
|
neurexophilin 1 |
| chr9_-_92435363 | 0.48 |
ENSRNOT00000093735
ENSRNOT00000022822 ENSRNOT00000093245 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr3_-_171134655 | 0.31 |
ENSRNOT00000028960
|
RGD1561252
|
similar to Ubiquitin carboxyl-terminal hydrolase isozyme L3 (Ubiquitin thiolesterase L3) |
| chr5_-_126911520 | 0.28 |
ENSRNOT00000091521
|
Dio1
|
deiodinase, iodothyronine, type I |
| chr7_+_70580198 | 0.23 |
ENSRNOT00000083472
ENSRNOT00000008941 |
Ddit3
|
DNA-damage inducible transcript 3 |
| chr5_+_116421894 | 0.13 |
ENSRNOT00000080577
ENSRNOT00000086628 ENSRNOT00000004017 |
Nfia
|
nuclear factor I/A |
| chr4_+_153385205 | 0.08 |
ENSRNOT00000016561
|
Bcl2l13
|
BCL2 like 13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Thra
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 27.3 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 3.9 | 11.6 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) multicellular organism lipid catabolic process(GO:0044240) |
| 3.7 | 11.0 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 3.6 | 10.9 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 2.9 | 11.6 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 2.5 | 7.6 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 2.3 | 27.5 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 2.0 | 16.0 | GO:0048840 | otolith development(GO:0048840) |
| 1.7 | 10.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 1.6 | 4.8 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 1.5 | 2.9 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.4 | 4.3 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 1.4 | 4.3 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 1.4 | 4.1 | GO:0021917 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 1.4 | 8.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 1.3 | 3.9 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 1.3 | 5.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) cytidine to uridine editing(GO:0016554) |
| 1.2 | 9.8 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.2 | 4.7 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 1.1 | 4.3 | GO:0038109 | Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.9 | 9.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.9 | 8.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.7 | 12.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.6 | 18.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.5 | 1.6 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.5 | 16.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.4 | 7.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.4 | 7.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.4 | 7.7 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.4 | 9.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.4 | 3.0 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.4 | 19.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.4 | 4.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.4 | 22.4 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.3 | 1.0 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.3 | 1.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.3 | 9.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.3 | 3.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.3 | 3.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.3 | 3.0 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.3 | 4.0 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 0.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 4.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 15.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 3.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 9.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 3.0 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.2 | 0.7 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.2 | 6.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.2 | 2.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 2.9 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 1.0 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 4.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.5 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.9 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 1.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 1.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 4.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 2.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 7.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 1.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.2 | GO:0051409 | response to nitrosative stress(GO:0051409) cellular response to nitrosative stress(GO:0071500) regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 4.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 6.4 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 1.9 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 5.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.8 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 1.4 | GO:0007601 | visual perception(GO:0007601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 38.9 | GO:0042627 | chylomicron(GO:0042627) |
| 2.2 | 10.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 2.1 | 8.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 1.6 | 19.2 | GO:0005861 | troponin complex(GO:0005861) |
| 1.4 | 4.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 1.3 | 16.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 1.1 | 7.7 | GO:1990357 | terminal web(GO:1990357) |
| 1.1 | 7.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 1.0 | 2.9 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.7 | 5.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.7 | 8.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.7 | 16.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.5 | 1.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.4 | 4.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 4.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 12.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 9.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 12.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 15.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 11.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 9.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 2.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 9.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 1.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 9.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 7.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 3.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 24.3 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 66.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 6.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 10.2 | GO:0044456 | synapse part(GO:0044456) |
| 0.0 | 1.2 | GO:0016363 | nuclear matrix(GO:0016363) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.1 | 27.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 4.3 | 29.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 4.0 | 16.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 3.6 | 10.9 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 3.2 | 19.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 2.1 | 8.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 2.0 | 9.8 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 2.0 | 11.8 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 1.9 | 11.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 1.9 | 15.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 1.5 | 7.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.0 | 4.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 1.0 | 4.8 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.9 | 8.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.6 | 3.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.6 | 7.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.5 | 9.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.4 | 3.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.4 | 11.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.4 | 4.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 10.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 27.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.4 | 4.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.4 | 9.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 4.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 6.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 4.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 4.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 2.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 12.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 1.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 11.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 7.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 4.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 3.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 12.6 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 9.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 5.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 7.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 7.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 5.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 10.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 16.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 3.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 4.3 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 1.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.0 | GO:0070851 | growth factor receptor binding(GO:0070851) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 15.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 42.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 6.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 4.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 26.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 9.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 4.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 27.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.6 | 27.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.5 | 19.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 12.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.5 | 7.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.4 | 11.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.4 | 4.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.4 | 7.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.4 | 9.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 9.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 9.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 3.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 4.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 9.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 30.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 3.0 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 6.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 5.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 4.3 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 3.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |