Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
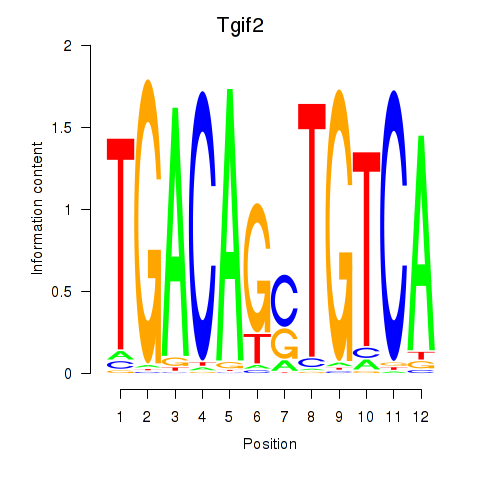
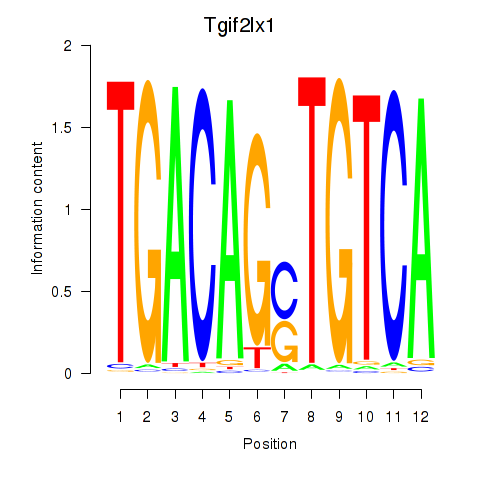
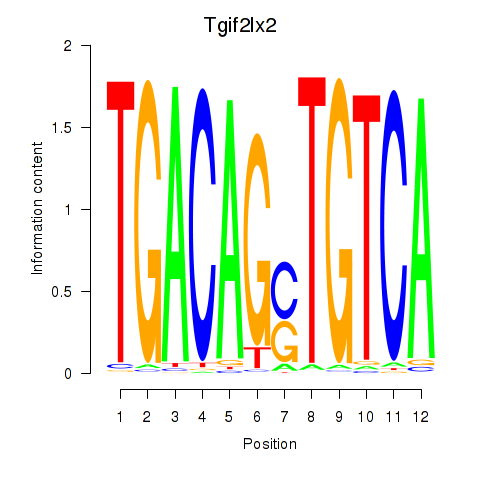
Results for Tgif2_Tgif2lx1_Tgif2lx2
Z-value: 0.94
Transcription factors associated with Tgif2_Tgif2lx1_Tgif2lx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tgif2
|
ENSRNOG00000042374 | TGFB-induced factor homeobox 2 |
|
Tgif2lx2
|
ENSRNOG00000029850 | TGFB-induced factor homeobox 2-like, X-linked 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tgif2lx2 | rn6_v1_chrX_+_90588124_90588124 | 0.17 | 2.7e-03 | Click! |
| Tgif2 | rn6_v1_chr3_+_152909189_152909189 | -0.12 | 2.7e-02 | Click! |
Activity profile of Tgif2_Tgif2lx1_Tgif2lx2 motif
Sorted Z-values of Tgif2_Tgif2lx1_Tgif2lx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_72219246 | 69.98 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr4_+_144382945 | 35.62 |
ENSRNOT00000007601
|
Cav3
|
caveolin 3 |
| chr1_+_80321585 | 35.34 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr7_-_29152442 | 33.21 |
ENSRNOT00000079774
|
Mybpc1
|
myosin binding protein C, slow type |
| chr5_+_154522119 | 31.71 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr9_-_43116521 | 25.51 |
ENSRNOT00000039437
|
Ankrd23
|
ankyrin repeat domain 23 |
| chr13_+_52889737 | 23.00 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr3_-_121822436 | 21.67 |
ENSRNOT00000039794
|
Ckap2l
|
cytoskeleton associated protein 2-like |
| chr10_+_59529785 | 20.29 |
ENSRNOT00000064840
ENSRNOT00000065181 |
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr16_+_75572070 | 15.96 |
ENSRNOT00000043486
|
Defb52
|
defensin beta 52 |
| chr2_-_88135410 | 15.93 |
ENSRNOT00000014180
|
Car3
|
carbonic anhydrase 3 |
| chr3_-_16753987 | 14.96 |
ENSRNOT00000091257
|
AABR07051548.1
|
|
| chr10_-_74413745 | 13.82 |
ENSRNOT00000038296
|
Prr11
|
proline rich 11 |
| chrX_-_138435391 | 13.56 |
ENSRNOT00000043258
|
Mbnl3
|
muscleblind-like splicing regulator 3 |
| chrX_+_136460215 | 12.76 |
ENSRNOT00000093538
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr1_-_145870912 | 12.36 |
ENSRNOT00000016289
|
Il16
|
interleukin 16 |
| chr5_+_58393603 | 11.67 |
ENSRNOT00000080082
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr14_-_80130139 | 11.56 |
ENSRNOT00000091652
ENSRNOT00000010482 |
Ablim2
|
actin binding LIM protein family, member 2 |
| chr14_-_34115273 | 11.51 |
ENSRNOT00000032156
|
Cep135
|
centrosomal protein 135 |
| chr3_+_18315320 | 11.07 |
ENSRNOT00000006954
|
AABR07051626.2
|
|
| chr3_+_19772056 | 10.54 |
ENSRNOT00000044455
|
AABR07051708.1
|
|
| chr2_+_40000313 | 9.94 |
ENSRNOT00000014270
|
Depdc1b
|
DEP domain containing 1B |
| chr1_-_100537377 | 9.78 |
ENSRNOT00000026599
|
Spib
|
Spi-B transcription factor |
| chr10_+_47412582 | 9.36 |
ENSRNOT00000003139
|
Tnfrsf13b
|
TNF receptor superfamily member 13B |
| chr8_-_39190457 | 9.09 |
ENSRNOT00000090161
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr8_-_48850671 | 9.06 |
ENSRNOT00000016580
|
Cxcr5
|
C-X-C motif chemokine receptor 5 |
| chr3_-_16750564 | 8.69 |
ENSRNOT00000084111
|
AABR07051548.1
|
|
| chr6_-_143702033 | 8.55 |
ENSRNOT00000051410
|
AABR07065886.1
|
|
| chr5_+_126670825 | 8.12 |
ENSRNOT00000012201
|
Cyb5rl
|
cytochrome b5 reductase-like |
| chr10_+_17486579 | 8.06 |
ENSRNOT00000080404
|
Stk10
|
serine/threonine kinase 10 |
| chr10_-_40489808 | 7.44 |
ENSRNOT00000037274
|
Slc36a3
|
solute carrier family 36, member 3 |
| chr4_+_145489869 | 7.44 |
ENSRNOT00000082618
|
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr4_-_60358562 | 7.06 |
ENSRNOT00000018001
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr6_-_138640187 | 7.03 |
ENSRNOT00000087983
|
AABR07065651.6
|
|
| chr6_+_139345486 | 6.52 |
ENSRNOT00000081540
|
AABR07065688.1
|
|
| chr1_+_140602542 | 6.47 |
ENSRNOT00000085570
|
Isg20
|
interferon stimulated exonuclease gene 20 |
| chr7_-_74938663 | 6.41 |
ENSRNOT00000014333
|
Fbxo43
|
F-box protein 43 |
| chr1_-_24604400 | 6.41 |
ENSRNOT00000081175
|
1700020N01Rik
|
RIKEN cDNA 1700020N01 gene |
| chr2_+_198655437 | 6.33 |
ENSRNOT00000028781
|
Hfe2
|
hemochromatosis type 2 (juvenile) |
| chr9_+_16530074 | 6.24 |
ENSRNOT00000091565
|
Ptcra
|
pre T-cell antigen receptor alpha |
| chr9_-_114152716 | 5.94 |
ENSRNOT00000044153
|
AABR07068650.1
|
|
| chr9_+_25304876 | 5.92 |
ENSRNOT00000065053
|
Tfap2d
|
transcription factor AP-2 delta |
| chr6_-_128727374 | 5.86 |
ENSRNOT00000082152
|
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr9_-_114282799 | 5.77 |
ENSRNOT00000090539
|
LOC102555328
|
uncharacterized LOC102555328 |
| chr3_+_72395218 | 5.76 |
ENSRNOT00000057616
|
Prg3
|
proteoglycan 3, pro eosinophil major basic protein 2 |
| chr16_-_19349080 | 5.69 |
ENSRNOT00000038494
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr4_-_164211819 | 5.60 |
ENSRNOT00000084796
|
LOC497796
|
hypothetical protein LOC497796 |
| chr15_+_5319916 | 5.51 |
ENSRNOT00000046644
|
LOC102546495
|
disks large homolog 5-like |
| chr7_-_27136652 | 5.51 |
ENSRNOT00000086905
|
Hcfc2
|
host cell factor C2 |
| chr6_-_125317593 | 5.50 |
ENSRNOT00000074142
|
LOC102553138
|
ovarian cancer G-protein coupled receptor 1-like |
| chr17_+_28504623 | 5.48 |
ENSRNOT00000021568
|
F13a1
|
coagulation factor XIII A1 chain |
| chr10_+_4945911 | 5.48 |
ENSRNOT00000003420
|
Prm1
|
protamine 1 |
| chr4_-_103646906 | 5.45 |
ENSRNOT00000047620
|
AABR07061078.1
|
|
| chr9_+_42871950 | 5.38 |
ENSRNOT00000089673
|
Arid5a
|
AT-rich interaction domain 5A |
| chr2_+_122368265 | 5.38 |
ENSRNOT00000078321
|
Atp11b
|
ATPase phospholipid transporting 11B (putative) |
| chr6_+_52265930 | 5.23 |
ENSRNOT00000087513
ENSRNOT00000067849 |
Sypl1
|
synaptophysin-like 1 |
| chr6_-_124876710 | 5.23 |
ENSRNOT00000073530
|
Gpr68
|
G protein-coupled receptor 68 |
| chr14_-_21709084 | 5.22 |
ENSRNOT00000087477
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chr10_+_56591292 | 5.21 |
ENSRNOT00000023379
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1 |
| chr4_-_130659697 | 5.20 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr9_-_16820983 | 5.20 |
ENSRNOT00000083886
|
Dmrtc2
|
DMRT-like family C2 |
| chr6_-_142676432 | 5.07 |
ENSRNOT00000074947
|
AABR07065815.2
|
|
| chr8_+_80662762 | 4.93 |
ENSRNOT00000057689
|
LOC100912027
|
60S ribosomal protein L27-like |
| chr7_+_120202601 | 4.87 |
ENSRNOT00000082862
|
AC096473.3
|
|
| chr1_+_140601791 | 4.80 |
ENSRNOT00000091588
|
Isg20
|
interferon stimulated exonuclease gene 20 |
| chr3_+_16590244 | 4.72 |
ENSRNOT00000073229
|
AABR07051535.1
|
|
| chr1_+_81736335 | 4.47 |
ENSRNOT00000027242
|
LOC100910371
|
doublesex- and mab-3-related transcription factor C2-like |
| chr12_+_25498198 | 4.45 |
ENSRNOT00000076916
|
Ncf1
|
neutrophil cytosolic factor 1 |
| chr3_-_72895740 | 4.44 |
ENSRNOT00000012568
|
Fads2l1
|
fatty acid desaturase 2-like 1 |
| chr2_+_123790915 | 4.38 |
ENSRNOT00000023282
|
Adad1
|
adenosine deaminase domain containing 1 |
| chr15_+_57221292 | 4.33 |
ENSRNOT00000014502
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr5_+_156439633 | 4.31 |
ENSRNOT00000090232
|
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr19_-_55423052 | 4.29 |
ENSRNOT00000019528
ENSRNOT00000079641 |
Galns
|
galactosamine (N-acetyl)-6-sulfatase |
| chr8_+_60760078 | 4.27 |
ENSRNOT00000063930
|
Pstpip1
|
proline-serine-threonine phosphatase-interacting protein 1 |
| chr5_+_155794229 | 4.27 |
ENSRNOT00000018127
|
LOC690206
|
hypothetical protein LOC690206 |
| chr4_+_79868442 | 4.20 |
ENSRNOT00000089973
|
Mpp6
|
membrane palmitoylated protein 6 |
| chr8_+_53411316 | 4.19 |
ENSRNOT00000011107
ENSRNOT00000086957 |
Tmprss5
|
transmembrane protease, serine 5 |
| chr4_-_68349273 | 4.18 |
ENSRNOT00000016251
|
Prss37
|
protease, serine, 37 |
| chr7_-_117259141 | 4.17 |
ENSRNOT00000040762
|
Plec
|
plectin |
| chr19_-_24875137 | 4.09 |
ENSRNOT00000006152
|
Adgre5
|
adhesion G protein-coupled receptor E5 |
| chr10_-_103590607 | 4.07 |
ENSRNOT00000034741
|
Cd300le
|
Cd300 molecule-like family member E |
| chr11_+_64472072 | 4.06 |
ENSRNOT00000042756
|
RGD1563835
|
similar to ribosomal protein L27 |
| chr7_-_117259791 | 4.03 |
ENSRNOT00000086550
|
Plec
|
plectin |
| chr18_+_3565166 | 4.01 |
ENSRNOT00000039377
|
Riok3
|
RIO kinase 3 |
| chr12_-_2843772 | 3.97 |
ENSRNOT00000061750
|
Cd209c
|
CD209c molecule |
| chr3_-_7203420 | 3.92 |
ENSRNOT00000015236
|
Gfi1b
|
growth factor independent 1B transcriptional repressor |
| chr12_+_37211316 | 3.85 |
ENSRNOT00000032739
|
Ccdc92
|
coiled-coil domain containing 92 |
| chr19_+_42097995 | 3.80 |
ENSRNOT00000020197
|
Hp
|
haptoglobin |
| chr10_+_110445797 | 3.80 |
ENSRNOT00000054920
|
Narf
|
nuclear prelamin A recognition factor |
| chr16_+_68586235 | 3.76 |
ENSRNOT00000039592
|
LOC103693984
|
uncharacterized LOC103693984 |
| chr6_-_143206772 | 3.75 |
ENSRNOT00000073713
|
AABR07065839.1
|
|
| chr19_-_23554594 | 3.74 |
ENSRNOT00000004590
|
Il15
|
interleukin 15 |
| chr2_+_248219428 | 3.69 |
ENSRNOT00000037181
|
LOC685067
|
similar to guanylate binding protein family, member 6 |
| chr9_+_10061978 | 3.53 |
ENSRNOT00000075180
|
Acer1
|
alkaline ceramidase 1 |
| chr2_-_27364906 | 3.50 |
ENSRNOT00000078639
|
Polk
|
DNA polymerase kappa |
| chr8_-_14373894 | 3.49 |
ENSRNOT00000011883
|
Mtnr1b
|
melatonin receptor 1B |
| chr1_-_48070536 | 3.49 |
ENSRNOT00000020070
|
Mas1l
|
MAS1 proto-oncogene like, G protein-coupled receptor |
| chr3_+_149144795 | 3.42 |
ENSRNOT00000015482
|
Dnmt3b
|
DNA methyltransferase 3 beta |
| chr2_+_210668403 | 3.37 |
ENSRNOT00000074806
|
Eps8l3
|
EPS8-like 3 |
| chr1_-_173180745 | 3.36 |
ENSRNOT00000043904
|
Olr203
|
olfactory receptor 203 |
| chr13_-_27653839 | 3.35 |
ENSRNOT00000071638
|
LOC100360575
|
signal-regulatory protein alpha-like |
| chrX_-_14783792 | 3.29 |
ENSRNOT00000087609
|
LOC691895
|
similar to ferritin, heavy polypeptide-like 17 |
| chr17_+_9639330 | 3.14 |
ENSRNOT00000018232
|
Dok3
|
docking protein 3 |
| chr20_+_4162055 | 3.13 |
ENSRNOT00000081236
|
Btnl3
|
butyrophilin-like 3 |
| chr18_-_11906827 | 3.08 |
ENSRNOT00000078207
|
Dsc1
|
desmocollin 1 |
| chr10_-_2229493 | 3.03 |
ENSRNOT00000047732
|
LOC688899
|
similar to 40S ribosomal protein S19 |
| chr14_+_39368530 | 3.03 |
ENSRNOT00000084367
|
Cox7b2
|
cytochrome c oxidase subunit 7B2 |
| chr6_+_110749705 | 3.01 |
ENSRNOT00000084348
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr20_-_2734296 | 3.01 |
ENSRNOT00000082285
|
Btnl5
|
butyrophilin-like 5 |
| chr7_-_122340943 | 3.00 |
ENSRNOT00000078345
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr1_+_212714179 | 3.00 |
ENSRNOT00000054880
ENSRNOT00000025680 |
Olr292
|
olfactory receptor 292 |
| chr4_-_132111079 | 2.96 |
ENSRNOT00000013719
|
Eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr17_-_22863966 | 2.93 |
ENSRNOT00000046157
|
Tmem170b
|
transmembrane protein 170B |
| chr5_+_33097654 | 2.89 |
ENSRNOT00000008087
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr10_+_103562725 | 2.88 |
ENSRNOT00000075199
|
AABR07030773.1
|
|
| chrX_-_77675487 | 2.85 |
ENSRNOT00000042799
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chrX_+_158351156 | 2.83 |
ENSRNOT00000080538
|
LOC100909732
|
protein FAM122B-like |
| chr14_-_37763712 | 2.81 |
ENSRNOT00000030610
|
Zar1
|
zygote arrest 1 |
| chr3_+_33641616 | 2.80 |
ENSRNOT00000051953
|
Epc2
|
enhancer of polycomb homolog 2 |
| chr7_+_143092511 | 2.79 |
ENSRNOT00000084223
|
RGD1305207
|
similar to RIKEN cDNA 1700011A15 |
| chr9_-_14706557 | 2.79 |
ENSRNOT00000048975
|
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr2_-_57600820 | 2.78 |
ENSRNOT00000083247
|
Nipbl
|
NIPBL, cohesin loading factor |
| chr1_-_266862842 | 2.76 |
ENSRNOT00000027496
|
LOC103693430
|
up-regulated during skeletal muscle growth protein 5 |
| chr2_+_80948658 | 2.74 |
ENSRNOT00000074689
|
Dnah5
|
dynein, axonemal, heavy chain 5 |
| chr17_+_15194262 | 2.73 |
ENSRNOT00000073070
|
AABR07072539.1
|
|
| chr3_+_21555341 | 2.72 |
ENSRNOT00000030332
|
AABR07051793.1
|
|
| chr15_+_24141651 | 2.70 |
ENSRNOT00000082304
|
Lgals3
|
galectin 3 |
| chr8_-_48762342 | 2.67 |
ENSRNOT00000049125
|
Foxr1
|
forkhead box R1 |
| chr2_-_246737997 | 2.65 |
ENSRNOT00000021719
|
Pdha2
|
pyruvate dehydrogenase (lipoamide) alpha 2 |
| chr15_+_30750093 | 2.64 |
ENSRNOT00000071830
|
AABR07017763.1
|
|
| chr4_+_95884743 | 2.63 |
ENSRNOT00000008585
|
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1` |
| chr8_-_76239995 | 2.61 |
ENSRNOT00000071321
|
LOC100912027
|
60S ribosomal protein L27-like |
| chr19_-_20508711 | 2.58 |
ENSRNOT00000032408
|
LOC680913
|
hypothetical protein LOC680913 |
| chr1_-_66645214 | 2.58 |
ENSRNOT00000084168
|
LOC100365062
|
eukaryotic translation initiation factor 3, subunit 6 48kDa-like |
| chr14_+_16924960 | 2.58 |
ENSRNOT00000003009
|
Ccdc158
|
coiled-coil domain containing 158 |
| chr17_+_25228437 | 2.54 |
ENSRNOT00000072904
|
AABR07027342.1
|
|
| chr8_-_105088401 | 2.51 |
ENSRNOT00000073068
|
LOC100365810
|
40S ribosomal protein S17-like |
| chr3_+_149466770 | 2.46 |
ENSRNOT00000065104
|
Bpifa6
|
BPI fold containing family A, member 6 |
| chr2_-_80293181 | 2.45 |
ENSRNOT00000016111
|
Otulin
|
OTU deubiquitinase with linear linkage specificity |
| chr11_-_47122095 | 2.44 |
ENSRNOT00000002194
|
Rpl24
|
ribosomal protein L24 |
| chr10_-_29196563 | 2.42 |
ENSRNOT00000005205
|
Fabp6
|
fatty acid binding protein 6 |
| chr2_+_219628695 | 2.36 |
ENSRNOT00000067324
|
Sass6
|
SAS-6 centriolar assembly protein |
| chr5_-_2803855 | 2.33 |
ENSRNOT00000009490
|
LOC297756
|
ribosomal protein S8-like |
| chr14_+_107767392 | 2.28 |
ENSRNOT00000012847
|
Cct4
|
chaperonin containing TCP1 subunit 4 |
| chrX_+_106230013 | 2.26 |
ENSRNOT00000067828
|
AABR07040617.1
|
|
| chr2_+_87418517 | 2.23 |
ENSRNOT00000048046
|
LOC100909879
|
tyrosine-protein phosphatase non-receptor type substrate 1-like |
| chr3_+_172195844 | 2.22 |
ENSRNOT00000034915
|
Npepl1
|
aminopeptidase-like 1 |
| chr20_-_7208292 | 2.21 |
ENSRNOT00000083169
|
Nudt3
|
nudix hydrolase 3 |
| chr14_-_14390699 | 2.19 |
ENSRNOT00000046639
|
Anxa3
|
annexin A3 |
| chr20_-_46215120 | 2.18 |
ENSRNOT00000000336
|
Smpd2
|
sphingomyelin phosphodiesterase 2 |
| chr1_+_248195797 | 2.17 |
ENSRNOT00000066891
|
LOC103690131
|
tumor protein D55-like |
| chr7_-_54823956 | 2.17 |
ENSRNOT00000073180
|
Glipr1l2
|
GLI pathogenesis-related 1 like 2 |
| chr13_+_48455923 | 2.12 |
ENSRNOT00000009280
|
Rab7b
|
Rab7b, member RAS oncogene family |
| chr3_+_149221377 | 2.07 |
ENSRNOT00000016220
ENSRNOT00000087752 |
Mapre1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr16_+_9907598 | 2.07 |
ENSRNOT00000027352
|
Ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chrX_-_123980357 | 2.03 |
ENSRNOT00000049435
|
Rhox8
|
reproductive homeobox 8 |
| chr1_-_124803363 | 2.03 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr7_+_70612103 | 2.01 |
ENSRNOT00000057833
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr2_-_35532285 | 1.99 |
ENSRNOT00000090178
|
LOC103691445
|
olfactory receptor 150-like |
| chr13_+_51218468 | 1.98 |
ENSRNOT00000033636
|
LOC289035
|
similar to UDP-N-acetylglucosamine:a-1,3-D-mannoside beta-1,4-N-acetylgluco |
| chr1_+_167539036 | 1.98 |
ENSRNOT00000093112
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chrX_-_23187341 | 1.96 |
ENSRNOT00000000180
|
Alas2
|
5'-aminolevulinate synthase 2 |
| chr11_-_31805728 | 1.95 |
ENSRNOT00000032162
|
Gart
|
phosphoribosylglycinamide formyltransferase |
| chr3_+_159095876 | 1.94 |
ENSRNOT00000074363
|
LOC103694889
|
seminal vesicle secretory protein 4 |
| chrX_+_63520991 | 1.91 |
ENSRNOT00000071590
|
Apoo
|
apolipoprotein O |
| chr3_-_149563476 | 1.88 |
ENSRNOT00000071869
|
AABR07054352.2
|
|
| chr2_+_155076464 | 1.88 |
ENSRNOT00000057619
|
LOC691033
|
similar to GTPase activating protein testicular GAP1 |
| chr6_-_26318568 | 1.80 |
ENSRNOT00000072707
|
LOC102556504
|
titin-like |
| chr13_+_77940454 | 1.80 |
ENSRNOT00000003460
|
Mrps14
|
mitochondrial ribosomal protein S14 |
| chr16_-_54628458 | 1.73 |
ENSRNOT00000042587
|
Adam24
|
ADAM metallopeptidase domain 24 |
| chr2_-_202470399 | 1.72 |
ENSRNOT00000026726
|
Wdr3
|
WD repeat domain 3 |
| chr20_-_2858205 | 1.71 |
ENSRNOT00000085662
|
Btnl8
|
butyrophilin-like 8 |
| chr10_+_43067299 | 1.67 |
ENSRNOT00000003447
|
Galnt10
|
polypeptide N-acetylgalactosaminyltransferase 10 |
| chr2_+_115225356 | 1.67 |
ENSRNOT00000073329
|
LOC100910086
|
olfactory receptor 149-like |
| chr17_-_89163113 | 1.66 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr2_+_115092263 | 1.66 |
ENSRNOT00000064455
|
LOC100910543
|
olfactory receptor 150-like |
| chr19_+_25123724 | 1.63 |
ENSRNOT00000007407
|
LOC686013
|
hypothetical protein LOC686013 |
| chr10_-_15928169 | 1.62 |
ENSRNOT00000028069
|
Nsg2
|
neuron specific gene family member 2 |
| chr1_-_173528943 | 1.62 |
ENSRNOT00000020393
|
Nlrp10
|
NLR family, pyrin domain containing 10 |
| chr1_-_7726401 | 1.61 |
ENSRNOT00000021524
|
Pex3
|
peroxisomal biogenesis factor 3 |
| chr16_+_6035926 | 1.59 |
ENSRNOT00000020284
|
Selenok
|
selenoprotein K |
| chr19_-_41433346 | 1.58 |
ENSRNOT00000022952
|
Cmtr2
|
cap methyltransferase 2 |
| chr5_+_126511350 | 1.54 |
ENSRNOT00000011408
|
Ssbp3
|
single stranded DNA binding protein 3 |
| chr20_-_5927070 | 1.53 |
ENSRNOT00000059264
|
Slc26a8
|
solute carrier family 26 member 8 |
| chr15_-_46297243 | 1.51 |
ENSRNOT00000088793
|
AABR07018191.1
|
|
| chr5_+_78267248 | 1.50 |
ENSRNOT00000019910
|
Prpf4
|
pre-mRNA processing factor 4 |
| chr8_-_130550388 | 1.50 |
ENSRNOT00000026355
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr4_-_179700130 | 1.49 |
ENSRNOT00000021306
|
Lmntd1
|
lamin tail domain containing 1 |
| chr7_-_130107437 | 1.48 |
ENSRNOT00000055865
|
Hdac10
|
histone deacetylase 10 |
| chr1_+_61314070 | 1.47 |
ENSRNOT00000077642
|
Zfp54
|
zinc finger protein 54 |
| chr1_+_167538744 | 1.46 |
ENSRNOT00000093070
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr5_+_173318479 | 1.42 |
ENSRNOT00000026725
|
Ints11
|
integrator complex subunit 11 |
| chr10_-_34439470 | 1.40 |
ENSRNOT00000072081
|
Btnl9
|
butyrophilin-like 9 |
| chr12_+_660011 | 1.38 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr17_+_66548818 | 1.34 |
ENSRNOT00000024361
|
Anlnl1
|
anillin, actin binding protein-like 1 |
| chr19_-_39646693 | 1.33 |
ENSRNOT00000019104
|
Psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr1_+_148862493 | 1.29 |
ENSRNOT00000073451
|
LOC100912097
|
olfactory receptor 10A5-like |
| chr4_-_1752548 | 1.28 |
ENSRNOT00000071677
|
LOC100912415
|
olfactory receptor 150-like |
| chr8_+_78872102 | 1.26 |
ENSRNOT00000090047
|
Zfp280d
|
zinc finger protein 280D |
| chr4_-_176679815 | 1.26 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tgif2_Tgif2lx1_Tgif2lx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 35.6 | GO:2001288 | detection of muscle stretch(GO:0035995) positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 5.3 | 31.7 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 3.8 | 23.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 3.8 | 11.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 3.7 | 33.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 2.8 | 25.5 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 1.9 | 5.8 | GO:0045575 | basophil activation(GO:0045575) |
| 1.7 | 5.2 | GO:1900114 | positive regulation of histone H3-K9 dimethylation(GO:1900111) positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 1.5 | 4.5 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 1.3 | 70.9 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 1.3 | 5.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 1.3 | 9.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 1.3 | 3.8 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 1.2 | 3.7 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 1.2 | 3.5 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) |
| 1.1 | 5.7 | GO:1904180 | negative regulation of membrane depolarization(GO:1904180) |
| 1.1 | 3.4 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 1.1 | 7.4 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.0 | 4.0 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 1.0 | 11.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.9 | 7.4 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.9 | 2.8 | GO:0035261 | external genitalia morphogenesis(GO:0035261) gall bladder development(GO:0061010) |
| 0.9 | 2.7 | GO:1903769 | negative regulation of cell proliferation in bone marrow(GO:1903769) |
| 0.9 | 3.5 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.9 | 4.3 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.8 | 2.5 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.7 | 5.9 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.7 | 2.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.7 | 2.1 | GO:1904528 | positive regulation of microtubule binding(GO:1904528) |
| 0.7 | 5.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.7 | 15.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.7 | 12.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.6 | 38.2 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.6 | 2.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.6 | 3.5 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.6 | 2.3 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.6 | 9.0 | GO:0042407 | cristae formation(GO:0042407) |
| 0.6 | 2.8 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.6 | 2.2 | GO:1901907 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.5 | 6.4 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.5 | 1.6 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.5 | 9.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.5 | 2.0 | GO:1901423 | response to benzene(GO:1901423) |
| 0.5 | 3.9 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.5 | 6.2 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.5 | 9.5 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.4 | 1.7 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.4 | 8.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 0.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 1.6 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.4 | 6.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.4 | 1.2 | GO:0033986 | response to methanol(GO:0033986) |
| 0.4 | 9.4 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.4 | 3.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.3 | 1.6 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.3 | 9.8 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.3 | 7.8 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 4.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 3.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 1.5 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.3 | 3.3 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 9.6 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.2 | 2.6 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.2 | 2.6 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 2.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 2.7 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.2 | 0.7 | GO:0090625 | siRNA loading onto RISC involved in RNA interference(GO:0035087) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.2 | 1.1 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 | 0.5 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.2 | 0.3 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.2 | 1.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 3.0 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.2 | 5.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 2.2 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.2 | 4.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 2.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 4.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.4 | GO:0036115 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.1 | 0.5 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 13.2 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.1 | 1.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 10.7 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.1 | 0.6 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.4 | GO:2000669 | neutrophil clearance(GO:0097350) negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.1 | 2.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.0 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 4.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 5.2 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 1.5 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.8 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 1.5 | GO:0019532 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.1 | 0.3 | GO:1904937 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.1 | 2.4 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 2.4 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 5.9 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 0.8 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 1.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 1.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.3 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 2.2 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 4.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.1 | GO:0031050 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.0 | 4.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 3.9 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 1.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 3.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 4.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.5 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 0.1 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.0 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 103.2 | GO:0031430 | M band(GO:0031430) |
| 1.9 | 35.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 1.3 | 9.0 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.1 | 6.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.0 | 3.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.9 | 3.4 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.8 | 5.9 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.7 | 2.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.7 | 43.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.6 | 2.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.5 | 9.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.5 | 9.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 5.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.5 | 1.5 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.5 | 2.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.5 | 3.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 4.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.5 | 2.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 4.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 8.2 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.4 | 25.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.3 | 2.6 | GO:0033290 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.3 | 2.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 1.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.3 | 5.2 | GO:0001741 | XY body(GO:0001741) |
| 0.3 | 2.6 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.3 | 21.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.2 | 11.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 3.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 5.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 1.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 2.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 3.0 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 4.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 15.4 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 2.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.7 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 2.7 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 0.9 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.6 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 3.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 32.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 3.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 3.7 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) |
| 0.1 | 5.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 16.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.8 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 4.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 5.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 9.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.8 | GO:0031902 | late endosome membrane(GO:0031902) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 35.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 4.5 | 35.6 | GO:0071253 | connexin binding(GO:0071253) |
| 3.9 | 70.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 2.6 | 33.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 2.3 | 11.3 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 2.0 | 15.9 | GO:0016151 | nickel cation binding(GO:0016151) |
| 1.8 | 25.5 | GO:0031432 | titin binding(GO:0031432) |
| 1.5 | 7.4 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 1.1 | 6.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 1.0 | 9.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.9 | 20.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.9 | 12.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.8 | 4.0 | GO:0089720 | caspase binding(GO:0089720) |
| 0.7 | 3.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.7 | 3.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.7 | 2.6 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.7 | 2.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.6 | 3.9 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.6 | 9.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.6 | 1.9 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.6 | 9.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.6 | 3.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.6 | 2.9 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.6 | 2.8 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.6 | 2.2 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.5 | 5.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 3.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.5 | 2.7 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.5 | 1.6 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.5 | 4.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.4 | 2.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.4 | 7.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.4 | 4.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.3 | 6.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 2.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 4.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.3 | 1.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.3 | 2.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 8.2 | GO:0030506 | structural constituent of muscle(GO:0008307) ankyrin binding(GO:0030506) |
| 0.2 | 2.9 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 3.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 9.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 0.9 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.2 | 3.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 5.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 4.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 1.5 | GO:0030621 | U6 snRNA binding(GO:0017070) U4 snRNA binding(GO:0030621) |
| 0.2 | 1.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 11.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.2 | 3.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 1.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 1.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 0.8 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 1.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 13.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 32.0 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 7.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 3.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 3.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 3.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 6.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 1.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.3 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.7 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 2.2 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 5.6 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 9.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 5.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 2.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.9 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 3.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 3.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 2.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.2 | GO:0001948 | glycoprotein binding(GO:0001948) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 31.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 1.5 | 35.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.5 | 36.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 9.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 5.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 7.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 6.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 4.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 21.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 3.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 2.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 31.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.9 | 16.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.9 | 33.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.7 | 7.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.5 | 22.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.4 | 4.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 8.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 1.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 2.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 2.9 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 2.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 3.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 13.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 11.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 9.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 1.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 2.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 22.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 26.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 6.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 7.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 4.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 9.1 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 3.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 0.8 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 1.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 4.0 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 1.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 2.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.3 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |