Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
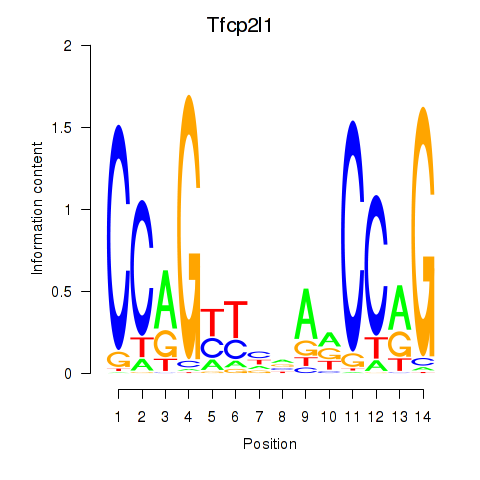
Results for Tfcp2l1
Z-value: 0.53
Transcription factors associated with Tfcp2l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfcp2l1
|
ENSRNOG00000002414 | transcription factor CP2-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfcp2l1 | rn6_v1_chr13_+_34610684_34610689 | 0.43 | 7.0e-16 | Click! |
Activity profile of Tfcp2l1 motif
Sorted Z-values of Tfcp2l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_105116916 | 16.18 |
ENSRNOT00000012373
|
Evpl
|
envoplakin |
| chr11_-_28527890 | 15.02 |
ENSRNOT00000002138
|
Cldn8
|
claudin 8 |
| chr8_-_50231357 | 14.48 |
ENSRNOT00000081737
|
Tagln
|
transgelin |
| chr13_+_47454591 | 14.17 |
ENSRNOT00000005791
|
LOC498222
|
similar to specifically androgen-regulated protein |
| chr2_-_204254699 | 13.34 |
ENSRNOT00000021487
|
Mab21l3
|
mab-21 like 3 |
| chr5_-_151824633 | 7.98 |
ENSRNOT00000043959
|
Sfn
|
stratifin |
| chr10_+_89251370 | 7.76 |
ENSRNOT00000076820
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr8_+_115151627 | 7.15 |
ENSRNOT00000064252
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr7_-_139483997 | 6.64 |
ENSRNOT00000086062
|
Col2a1
|
collagen type II alpha 1 chain |
| chr2_-_195908037 | 6.42 |
ENSRNOT00000079776
|
Tuft1
|
tuftelin 1 |
| chr20_+_5049496 | 6.42 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr3_+_113319456 | 6.36 |
ENSRNOT00000051354
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr1_+_264504591 | 5.78 |
ENSRNOT00000050076
|
Pax2
|
paired box 2 |
| chr13_-_45318822 | 5.61 |
ENSRNOT00000091514
ENSRNOT00000005143 |
Cxcr4
|
C-X-C motif chemokine receptor 4 |
| chr5_-_153924896 | 5.16 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr10_+_63677396 | 4.98 |
ENSRNOT00000005100
|
Slc43a2
|
solute carrier family 43 member 2 |
| chr8_+_85503224 | 4.98 |
ENSRNOT00000012348
|
Gsta4
|
glutathione S-transferase alpha 4 |
| chr4_-_116278615 | 4.27 |
ENSRNOT00000020505
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr6_+_23757225 | 4.22 |
ENSRNOT00000005553
|
LOC298795
|
similar to 14-3-3 protein sigma |
| chr2_+_200572502 | 4.12 |
ENSRNOT00000074666
|
Zfp697
|
zinc finger protein 697 |
| chr1_-_141481315 | 4.03 |
ENSRNOT00000020229
|
Pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr7_-_143228060 | 3.97 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr11_+_39482408 | 3.79 |
ENSRNOT00000075126
|
Hmgn1
|
high mobility group nucleosome binding domain 1 |
| chr17_+_30965942 | 3.54 |
ENSRNOT00000060143
|
Pxdc1
|
PX domain containing 1 |
| chr3_+_170550314 | 3.48 |
ENSRNOT00000006991
|
Tfap2c
|
transcription factor AP-2 gamma |
| chr1_+_225129097 | 3.28 |
ENSRNOT00000026938
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr2_+_196334626 | 3.25 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr8_-_21661259 | 3.22 |
ENSRNOT00000068308
|
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr1_-_199037267 | 3.16 |
ENSRNOT00000078779
|
Ccdc189
|
coiled-coil domain containing 189 |
| chr11_-_36479868 | 3.15 |
ENSRNOT00000075762
|
LOC100911295
|
non-histone chromosomal protein HMG-14-like |
| chr9_+_81518584 | 3.13 |
ENSRNOT00000084309
|
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr9_+_81518176 | 3.07 |
ENSRNOT00000078317
ENSRNOT00000019265 ENSRNOT00000088246 ENSRNOT00000084682 |
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr16_-_36080191 | 3.02 |
ENSRNOT00000017635
|
Hmgb2l1
|
high mobility group box 2-like 1 |
| chr3_+_176691329 | 2.98 |
ENSRNOT00000017319
|
Ppdpf
|
pancreatic progenitor cell differentiation and proliferation factor |
| chr9_+_81672758 | 2.91 |
ENSRNOT00000020646
|
Ctdsp1
|
CTD small phosphatase 1 |
| chr1_+_199323628 | 2.85 |
ENSRNOT00000036187
|
Zfp646
|
zinc finger protein 646 |
| chr3_+_161236898 | 2.62 |
ENSRNOT00000082303
ENSRNOT00000020323 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr7_-_61798729 | 2.48 |
ENSRNOT00000010283
|
Dyrk2
|
dual specificity tyrosine phosphorylation regulated kinase 2 |
| chr10_-_57837602 | 2.41 |
ENSRNOT00000075185
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr7_+_79138925 | 2.40 |
ENSRNOT00000047511
|
AABR07057586.1
|
|
| chr1_+_44446765 | 2.35 |
ENSRNOT00000030132
|
Cldn20
|
claudin 20 |
| chr5_-_58124681 | 2.23 |
ENSRNOT00000019795
|
Sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr11_-_14741563 | 2.23 |
ENSRNOT00000002152
|
Nrip1
|
nuclear receptor interacting protein 1 |
| chr13_+_52854031 | 2.19 |
ENSRNOT00000013738
ENSRNOT00000086004 |
Tmem9
|
transmembrane protein 9 |
| chr20_+_3553455 | 2.13 |
ENSRNOT00000090080
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr10_-_64862268 | 2.06 |
ENSRNOT00000056234
|
Phf12
|
PHD finger protein 12 |
| chr6_-_9459188 | 2.03 |
ENSRNOT00000019894
|
Srbd1
|
S1 RNA binding domain 1 |
| chr11_+_36075709 | 2.01 |
ENSRNOT00000002247
|
Ets2
|
ETS proto-oncogene 2, transcription factor |
| chr4_+_145427367 | 1.94 |
ENSRNOT00000037788
|
Il17rc
|
interleukin 17 receptor C |
| chr3_+_120726906 | 1.93 |
ENSRNOT00000051069
|
Bcl2l11
|
BCL2 like 11 |
| chr10_-_92476109 | 1.87 |
ENSRNOT00000089029
|
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr9_-_20195566 | 1.83 |
ENSRNOT00000015223
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr10_-_90265017 | 1.78 |
ENSRNOT00000064283
ENSRNOT00000048418 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr8_+_116754178 | 1.77 |
ENSRNOT00000068295
ENSRNOT00000084429 |
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr6_+_42852683 | 1.76 |
ENSRNOT00000079185
|
Odc1
|
ornithine decarboxylase 1 |
| chr1_-_225128740 | 1.74 |
ENSRNOT00000026897
|
Rom1
|
retinal outer segment membrane protein 1 |
| chr10_+_59893067 | 1.62 |
ENSRNOT00000056443
|
Spata22
|
spermatogenesis associated 22 |
| chr10_+_87759769 | 1.60 |
ENSRNOT00000017378
ENSRNOT00000046526 |
Krtap9-1
|
keratin associated protein 9-1 |
| chr15_-_33285779 | 1.60 |
ENSRNOT00000036150
|
Cdh24
|
cadherin 24 |
| chr2_+_199199845 | 1.56 |
ENSRNOT00000020932
|
LOC100909441
|
tubulin alpha-1C chain-like |
| chr14_-_91904433 | 1.53 |
ENSRNOT00000005857
|
Fignl1
|
fidgetin-like 1 |
| chr19_+_32188267 | 1.45 |
ENSRNOT00000025079
|
Smad1
|
SMAD family member 1 |
| chr10_+_106712127 | 1.27 |
ENSRNOT00000040629
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr5_-_164747083 | 1.21 |
ENSRNOT00000010433
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr11_+_74050166 | 1.20 |
ENSRNOT00000002348
|
Lrrc15
|
leucine rich repeat containing 15 |
| chr3_-_8659102 | 1.08 |
ENSRNOT00000050908
|
Zdhhc12
|
zinc finger, DHHC-type containing 12 |
| chr8_+_71719563 | 1.05 |
ENSRNOT00000022828
|
Ppib
|
peptidylprolyl isomerase B |
| chr20_-_12820466 | 1.03 |
ENSRNOT00000001699
|
Ftcd
|
formimidoyltransferase cyclodeaminase |
| chr5_-_152198813 | 1.01 |
ENSRNOT00000082953
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr1_-_164205222 | 0.99 |
ENSRNOT00000035774
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr14_+_108007724 | 0.87 |
ENSRNOT00000014062
|
Xpo1
|
exportin 1 |
| chr9_+_47281961 | 0.85 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr11_-_28777603 | 0.81 |
ENSRNOT00000002132
|
Krtap14
|
keratin associated protein 14 |
| chr11_-_82860977 | 0.79 |
ENSRNOT00000002382
|
Polr2h
|
RNA polymerase II subunit H |
| chr15_+_34552410 | 0.78 |
ENSRNOT00000027802
|
Khnyn
|
KH and NYN domain containing |
| chr10_+_66509107 | 0.73 |
ENSRNOT00000017230
|
RGD1565317
|
similar to ubiquitin-like/S30 ribosomal fusion protein |
| chr10_-_87510629 | 0.71 |
ENSRNOT00000084197
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr19_+_57650163 | 0.65 |
ENSRNOT00000038257
ENSRNOT00000083572 |
Sprtn
|
SprT-like N-terminal domain |
| chr10_-_110555629 | 0.64 |
ENSRNOT00000054918
ENSRNOT00000088474 |
Wdr45b
|
WD repeat domain 45B |
| chr3_+_150055749 | 0.63 |
ENSRNOT00000055335
|
Actl10
|
actin-like 10 |
| chr16_-_32421005 | 0.62 |
ENSRNOT00000082712
|
Nek1
|
NIMA-related kinase 1 |
| chr6_+_86131242 | 0.55 |
ENSRNOT00000039337
|
Fau
|
FAU, ubiquitin like and ribosomal protein S30 fusion |
| chr10_-_110101872 | 0.53 |
ENSRNOT00000071893
|
Ccdc57
|
coiled-coil domain containing 57 |
| chr10_+_87788458 | 0.53 |
ENSRNOT00000042020
|
LOC100910814
|
keratin-associated protein 9-1-like |
| chr15_-_56970365 | 0.45 |
ENSRNOT00000047192
|
Lrch1
|
leucine rich repeats and calponin homology domain containing 1 |
| chr4_+_130332076 | 0.43 |
ENSRNOT00000083127
|
Mitf
|
melanogenesis associated transcription factor |
| chr7_-_114590119 | 0.40 |
ENSRNOT00000079599
|
Ptk2
|
protein tyrosine kinase 2 |
| chr1_-_65701867 | 0.35 |
ENSRNOT00000006118
|
Zfp128
|
zinc finger protein 128 |
| chr6_-_142353308 | 0.29 |
ENSRNOT00000066416
|
AABR07065814.2
|
|
| chr6_-_125812517 | 0.24 |
ENSRNOT00000007061
|
Trip11
|
thyroid hormone receptor interactor 11 |
| chr10_+_87832743 | 0.24 |
ENSRNOT00000055286
|
Krtap31-1
|
keratin associated protein 31-1 |
| chr20_+_3555135 | 0.16 |
ENSRNOT00000085380
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr10_-_87510360 | 0.00 |
ENSRNOT00000077126
|
Krtap1-3
|
keratin associated protein 1-3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfcp2l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 1.6 | 6.2 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 1.4 | 4.3 | GO:1902809 | cornification(GO:0070268) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 1.4 | 5.6 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) response to ultrasound(GO:1990478) |
| 1.3 | 4.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 1.3 | 8.0 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 1.3 | 3.8 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 1.1 | 6.6 | GO:0071306 | cellular response to vitamin E(GO:0071306) |
| 0.7 | 2.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.7 | 2.6 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.6 | 1.8 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.6 | 3.5 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.5 | 2.0 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.5 | 6.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.5 | 1.9 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.4 | 7.8 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.4 | 1.2 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.4 | 16.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.4 | 2.5 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.4 | 1.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 1.0 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.3 | 5.2 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.3 | 1.0 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 0.3 | 1.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 5.0 | GO:0009635 | response to herbicide(GO:0009635) |
| 0.3 | 1.8 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.3 | 3.0 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 2.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 1.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 1.6 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.2 | 2.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 1.8 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.2 | 1.2 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 1.9 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 5.0 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 2.4 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 | 1.9 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.9 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.4 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 1.7 | GO:0061298 | camera-type eye photoreceptor cell differentiation(GO:0060219) retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.9 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 6.4 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.1 | 1.5 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 1.0 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.7 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 1.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.0 | 4.0 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 14.5 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.0 | 2.9 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.0 | 1.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.2 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.6 | 15.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.6 | 16.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.6 | 6.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 2.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.3 | 1.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 4.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 1.0 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 5.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.9 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 2.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 2.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 1.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 13.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 2.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 6.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 11.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 2.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 30.8 | GO:0070062 | extracellular exosome(GO:0070062) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 1.4 | 4.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 1.1 | 6.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.1 | 17.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 1.0 | 6.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.9 | 5.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.9 | 6.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.8 | 3.0 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.7 | 7.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.6 | 5.8 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.5 | 2.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.4 | 1.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.4 | 2.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.4 | 1.8 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.3 | 2.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 5.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.3 | 1.8 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.3 | 3.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 8.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 2.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 1.9 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 1.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.2 | 4.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 1.4 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.2 | 1.8 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 1.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 5.0 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 2.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 14.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 1.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 8.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 5.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 3.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 2.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 20.8 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.0 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 3.5 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 3.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 19.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 6.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 7.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 2.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 20.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 1.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 3.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 3.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.5 | 17.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 5.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 2.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.2 | 6.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 4.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 5.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.9 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 4.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |