Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
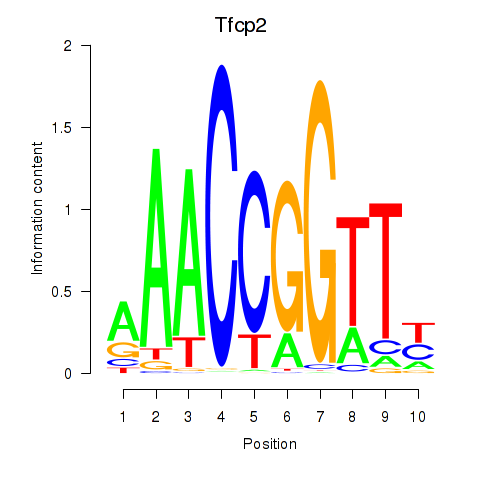
Results for Tfcp2
Z-value: 0.81
Transcription factors associated with Tfcp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfcp2
|
ENSRNOG00000032395 | transcription factor CP2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfcp2 | rn6_v1_chr7_-_142180794_142180794 | 0.55 | 6.7e-27 | Click! |
Activity profile of Tfcp2 motif
Sorted Z-values of Tfcp2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_133896141 | 28.67 |
ENSRNOT00000011434
|
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr19_-_11308740 | 22.80 |
ENSRNOT00000067391
|
Mt2A
|
metallothionein 2A |
| chr2_-_225107283 | 21.24 |
ENSRNOT00000055711
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr9_+_27402381 | 17.59 |
ENSRNOT00000077372
|
Gsta3
|
glutathione S-transferase alpha 3 |
| chr7_-_145154131 | 15.99 |
ENSRNOT00000055271
|
Ppp1r1a
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr20_+_20236151 | 15.02 |
ENSRNOT00000079630
|
Ank3
|
ankyrin 3 |
| chr5_-_33892462 | 14.80 |
ENSRNOT00000009334
|
Atp6v0d2
|
ATPase H+ transporting V0 subunit D2 |
| chr16_-_21348391 | 13.65 |
ENSRNOT00000083537
|
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr5_+_149077412 | 13.59 |
ENSRNOT00000014666
|
Matn1
|
matrilin 1, cartilage matrix protein |
| chr4_-_148845267 | 13.57 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr15_+_38341089 | 13.22 |
ENSRNOT00000015367
|
Fgf9
|
fibroblast growth factor 9 |
| chr10_+_3338808 | 12.59 |
ENSRNOT00000004384
ENSRNOT00000093444 |
Mpv17l
|
MPV17 mitochondrial inner membrane protein like |
| chr9_-_75528644 | 11.79 |
ENSRNOT00000019283
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chr20_+_48335540 | 11.77 |
ENSRNOT00000000352
|
Cd24
|
CD24 molecule |
| chr8_+_13796021 | 11.75 |
ENSRNOT00000013927
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr10_+_56228043 | 11.28 |
ENSRNOT00000015769
|
Sat2
|
spermidine/spermine N1-acetyltransferase family member 2 |
| chr20_-_19637958 | 10.51 |
ENSRNOT00000074657
|
Slc16a9
|
solute carrier family 16 (monocarboxylic acid transporters), member 9 |
| chr3_+_56802714 | 10.27 |
ENSRNOT00000077551
|
Erich2
|
glutamate-rich 2 |
| chr7_+_144623555 | 10.25 |
ENSRNOT00000022217
|
Hoxc6
|
homeo box C6 |
| chr5_-_158439078 | 9.93 |
ENSRNOT00000025517
|
Klhdc7a
|
kelch domain containing 7A |
| chr2_+_179952227 | 9.66 |
ENSRNOT00000015081
|
Pdgfc
|
platelet derived growth factor C |
| chr17_-_43504604 | 9.47 |
ENSRNOT00000083829
ENSRNOT00000066313 |
Slc17a1
|
solute carrier family 17 member 1 |
| chr7_-_126461658 | 9.36 |
ENSRNOT00000081032
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr16_-_21338771 | 9.32 |
ENSRNOT00000014265
|
Pbx4
|
PBX homeobox 4 |
| chr20_-_33323367 | 8.68 |
ENSRNOT00000080444
|
Ros1
|
ROS proto-oncogene 1 , receptor tyrosine kinase |
| chr4_-_82177420 | 8.56 |
ENSRNOT00000074452
|
LOC100909604
|
homeobox protein Hox-A6-like |
| chr6_+_50528823 | 8.43 |
ENSRNOT00000008321
|
Lamb1
|
laminin subunit beta 1 |
| chr1_+_105285419 | 8.43 |
ENSRNOT00000089693
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr4_+_163267450 | 8.24 |
ENSRNOT00000079337
|
Tmem52b
|
transmembrane protein 52B |
| chr4_-_82263117 | 8.17 |
ENSRNOT00000008542
|
LOC100909604
|
homeobox protein Hox-A6-like |
| chr17_+_30617382 | 8.06 |
ENSRNOT00000048923
|
Eci2
|
enoyl-CoA delta isomerase 2 |
| chrX_-_157172068 | 8.02 |
ENSRNOT00000087962
|
Dusp9
|
dual specificity phosphatase 9 |
| chr20_-_44220702 | 7.97 |
ENSRNOT00000036853
|
Fam229b
|
family with sequence similarity 229, member B |
| chr17_-_69404323 | 7.92 |
ENSRNOT00000051342
ENSRNOT00000066282 |
Akr1c2
|
aldo-keto reductase family 1, member C2 |
| chr20_-_33322966 | 7.88 |
ENSRNOT00000000459
|
Ros1
|
ROS proto-oncogene 1 , receptor tyrosine kinase |
| chr3_-_64095120 | 7.64 |
ENSRNOT00000016837
|
Sestd1
|
SEC14 and spectrin domain containing 1 |
| chr2_+_185393441 | 7.14 |
ENSRNOT00000015802
|
Sh3d19
|
SH3 domain containing 19 |
| chr1_+_105284753 | 7.12 |
ENSRNOT00000041950
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr17_-_32158538 | 7.04 |
ENSRNOT00000024141
|
Nqo2
|
NAD(P)H quinone dehydrogenase 2 |
| chr19_-_601469 | 7.03 |
ENSRNOT00000016462
|
Pdp2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr2_+_60169517 | 7.01 |
ENSRNOT00000080974
|
Prlr
|
prolactin receptor |
| chr14_-_20920286 | 6.76 |
ENSRNOT00000004391
|
Slc4a4
|
solute carrier family 4 member 4 |
| chr4_-_16669368 | 6.74 |
ENSRNOT00000007608
|
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr10_-_36716601 | 6.52 |
ENSRNOT00000038838
|
LOC497899
|
similar to hypothetical protein 4930503F14 |
| chr8_+_117268337 | 6.41 |
ENSRNOT00000072098
ENSRNOT00000083578 |
Lamb2
|
laminin subunit beta 2 |
| chr4_+_66090681 | 6.30 |
ENSRNOT00000078066
|
Ttc26
|
tetratricopeptide repeat domain 26 |
| chr4_-_117126822 | 6.23 |
ENSRNOT00000086720
|
Rab11fip5
|
RAB11 family interacting protein 5 |
| chr2_+_186980992 | 6.14 |
ENSRNOT00000020717
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr3_-_149905944 | 5.91 |
ENSRNOT00000021715
|
Snta1
|
syntrophin, alpha 1 |
| chr7_-_125795980 | 5.90 |
ENSRNOT00000017420
|
Arhgap8
|
Rho GTPase activating protein 8 |
| chr6_+_34028936 | 5.71 |
ENSRNOT00000085797
|
Laptm4a
|
lysosomal protein transmembrane 4 alpha |
| chrX_+_110789269 | 5.70 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr7_+_130296897 | 5.61 |
ENSRNOT00000044854
|
Adm2
|
adrenomedullin 2 |
| chr14_+_9226125 | 5.58 |
ENSRNOT00000088047
|
Wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr20_-_4879779 | 5.56 |
ENSRNOT00000081924
|
Hspa1b
|
heat shock protein family A (Hsp70) member 1B |
| chr11_+_33812662 | 5.37 |
ENSRNOT00000085961
|
LOC102556347
|
carbonyl reductase [NADPH] 1-like |
| chr7_-_142062870 | 5.29 |
ENSRNOT00000026531
|
Slc11a2
|
solute carrier family 11 member 2 |
| chr11_+_33812989 | 5.21 |
ENSRNOT00000042283
ENSRNOT00000075985 |
Cbr1
|
carbonyl reductase 1 |
| chr11_-_36479868 | 5.14 |
ENSRNOT00000075762
|
LOC100911295
|
non-histone chromosomal protein HMG-14-like |
| chr2_-_88135410 | 5.12 |
ENSRNOT00000014180
|
Car3
|
carbonic anhydrase 3 |
| chr15_+_32894938 | 5.11 |
ENSRNOT00000012837
|
Abhd4
|
abhydrolase domain containing 4 |
| chr2_+_186980793 | 5.10 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr1_+_166433109 | 5.08 |
ENSRNOT00000026428
|
Stard10
|
StAR-related lipid transfer domain containing 10 |
| chr9_+_61655963 | 4.95 |
ENSRNOT00000040461
|
Coq10b
|
coenzyme Q10B |
| chr2_+_187697523 | 4.86 |
ENSRNOT00000026153
|
Glmp
|
glycosylated lysosomal membrane protein |
| chr7_+_126028350 | 4.84 |
ENSRNOT00000042412
|
Ribc2
|
RIB43A domain with coiled-coils 2 |
| chr8_-_116993193 | 4.78 |
ENSRNOT00000026327
|
Dag1
|
dystroglycan 1 |
| chr1_+_199196059 | 4.73 |
ENSRNOT00000090428
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr2_+_45668969 | 4.70 |
ENSRNOT00000014761
ENSRNOT00000071353 |
Arl15
|
ADP-ribosylation factor like GTPase 15 |
| chr10_+_63677396 | 4.67 |
ENSRNOT00000005100
|
Slc43a2
|
solute carrier family 43 member 2 |
| chr2_+_225570721 | 4.56 |
ENSRNOT00000017076
|
Arhgap29
|
Rho GTPase activating protein 29 |
| chr20_+_5050327 | 4.54 |
ENSRNOT00000083353
|
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr1_+_214009784 | 4.52 |
ENSRNOT00000026079
|
LOC100911730
|
CD151 antigen-like |
| chr18_+_30885789 | 4.44 |
ENSRNOT00000044434
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr1_+_214446659 | 4.39 |
ENSRNOT00000072434
|
Cd151
|
CD151 molecule (Raph blood group) |
| chr20_+_5049496 | 4.39 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr5_+_4982348 | 4.36 |
ENSRNOT00000010369
|
Lactb2
|
lactamase, beta 2 |
| chr1_+_214009986 | 4.27 |
ENSRNOT00000089241
|
LOC100911730
|
CD151 antigen-like |
| chr4_-_82263302 | 4.25 |
ENSRNOT00000082933
|
LOC100909604
|
homeobox protein Hox-A6-like |
| chr6_-_8344574 | 4.21 |
ENSRNOT00000009660
|
Prepl
|
prolyl endopeptidase-like |
| chr9_+_65110330 | 4.12 |
ENSRNOT00000033068
ENSRNOT00000092868 |
Aox4
|
aldehyde oxidase 4 |
| chr10_+_92018562 | 4.07 |
ENSRNOT00000006483
|
Arf2
|
ADP-ribosylation factor 2 |
| chrX_-_104932508 | 4.05 |
ENSRNOT00000075325
|
Nox1
|
NADPH oxidase 1 |
| chr19_+_52217984 | 4.04 |
ENSRNOT00000079580
|
Dnaaf1
|
dynein, axonemal, assembly factor 1 |
| chrX_-_72133692 | 4.00 |
ENSRNOT00000004263
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr4_-_73174179 | 3.98 |
ENSRNOT00000048203
|
Tpk1
|
thiamin pyrophosphokinase 1 |
| chr1_-_81127059 | 3.95 |
ENSRNOT00000026252
|
Zfp94
|
zinc finger protein 94 |
| chr11_+_39482408 | 3.91 |
ENSRNOT00000075126
|
Hmgn1
|
high mobility group nucleosome binding domain 1 |
| chr7_+_97760134 | 3.83 |
ENSRNOT00000086792
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr7_-_126028279 | 3.73 |
ENSRNOT00000044883
|
Smc1b
|
structural maintenance of chromosomes 1B |
| chr6_-_102047758 | 3.73 |
ENSRNOT00000012101
|
Atp6v1d
|
ATPase H+ transporting V1 subunit D |
| chr19_-_41161765 | 3.69 |
ENSRNOT00000023117
|
Hydin
|
Hydin, axonemal central pair apparatus protein |
| chrX_+_118742313 | 3.68 |
ENSRNOT00000045110
|
AABR07041078.1
|
|
| chr1_-_250951697 | 3.67 |
ENSRNOT00000054761
|
Sgms1
|
sphingomyelin synthase 1 |
| chr1_-_220938814 | 3.66 |
ENSRNOT00000028081
|
Ovol1
|
ovo like transcriptional repressor 1 |
| chr5_+_157434481 | 3.64 |
ENSRNOT00000088556
|
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr3_-_45169118 | 3.63 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr11_+_61531416 | 3.63 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr2_-_195678848 | 3.62 |
ENSRNOT00000028303
ENSRNOT00000075569 |
Oaz3
|
ornithine decarboxylase antizyme 3 |
| chr2_-_210473761 | 3.59 |
ENSRNOT00000066501
|
Ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr13_-_67206688 | 3.50 |
ENSRNOT00000003630
ENSRNOT00000090693 |
Pla2g4a
|
phospholipase A2 group IVA |
| chr10_+_37594578 | 3.48 |
ENSRNOT00000007676
|
Skp1
|
S-phase kinase-associated protein 1 |
| chr18_+_29960072 | 3.46 |
ENSRNOT00000071366
|
AC103179.1
|
|
| chr2_-_144467912 | 3.38 |
ENSRNOT00000040002
|
Ccna1
|
cyclin A1 |
| chr11_-_31103520 | 3.38 |
ENSRNOT00000030279
|
RGD1306954
|
similar to RIKEN cDNA 1110004E09 |
| chr4_+_168832910 | 3.34 |
ENSRNOT00000011134
|
Gprc5a
|
G protein-coupled receptor, class C, group 5, member A |
| chr3_+_11554457 | 3.26 |
ENSRNOT00000073087
|
Fam102a
|
family with sequence similarity 102, member A |
| chr6_-_8344897 | 3.19 |
ENSRNOT00000082353
|
Prepl
|
prolyl endopeptidase-like |
| chr5_-_79570073 | 3.13 |
ENSRNOT00000011845
|
Tnfsf15
|
tumor necrosis factor superfamily member 15 |
| chr12_-_29958050 | 3.13 |
ENSRNOT00000058725
|
Tmem248
|
transmembrane protein 248 |
| chr19_+_37330930 | 3.12 |
ENSRNOT00000022439
|
Plekhg4
|
pleckstrin homology and RhoGEF domain containing G4 |
| chr3_+_103747654 | 2.91 |
ENSRNOT00000006887
|
Nop10
|
NOP10 ribonucleoprotein |
| chr16_+_27399467 | 2.91 |
ENSRNOT00000065642
|
Tll1
|
tolloid-like 1 |
| chr15_+_108453147 | 2.90 |
ENSRNOT00000018486
|
Timm8a2
|
translocase of inner mitochondrial membrane 8A2 |
| chr9_+_88110731 | 2.88 |
ENSRNOT00000088677
|
Rhbdd1
|
rhomboid domain containing 1 |
| chr9_-_71445739 | 2.85 |
ENSRNOT00000019698
|
Fzd5
|
frizzled class receptor 5 |
| chr7_+_3630950 | 2.75 |
ENSRNOT00000074535
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr1_-_280233755 | 2.72 |
ENSRNOT00000064463
|
Shtn1
|
shootin 1 |
| chr5_-_50193571 | 2.71 |
ENSRNOT00000051243
|
Cfap206
|
cilia and flagella associated protein 206 |
| chr10_+_105500290 | 2.70 |
ENSRNOT00000079080
ENSRNOT00000083593 |
Sphk1
|
sphingosine kinase 1 |
| chr5_+_146933592 | 2.66 |
ENSRNOT00000007851
|
A3galt2
|
alpha 1,3-galactosyltransferase 2 |
| chr6_+_43234526 | 2.65 |
ENSRNOT00000086808
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr1_+_91663736 | 2.62 |
ENSRNOT00000089587
ENSRNOT00000031236 |
Cep89
|
centrosomal protein 89 |
| chr1_+_98627372 | 2.60 |
ENSRNOT00000030370
|
Dzf17
|
zinc finger protein 17 |
| chrX_+_123404518 | 2.60 |
ENSRNOT00000015085
|
Slc25a5
|
solute carrier family 25 member 5 |
| chr1_-_188097374 | 2.56 |
ENSRNOT00000092246
|
Syt17
|
synaptotagmin 17 |
| chr1_-_71710374 | 2.53 |
ENSRNOT00000078556
ENSRNOT00000046152 |
Nlrp4
|
NLR family, pyrin domain containing 4 |
| chrX_-_143558521 | 2.51 |
ENSRNOT00000056598
|
LOC688842
|
hypothetical protein LOC688842 |
| chr8_+_21611319 | 2.48 |
ENSRNOT00000068461
|
Zfp846
|
zinc finger protein 846 |
| chr4_-_98735346 | 2.45 |
ENSRNOT00000008473
|
Tex37
|
testis expressed 37 |
| chr1_+_162419262 | 2.42 |
ENSRNOT00000016654
|
Kctd14
|
potassium channel tetramerization domain containing 14 |
| chr3_-_162872831 | 2.38 |
ENSRNOT00000008478
|
Sulf2
|
sulfatase 2 |
| chr1_+_264796812 | 2.37 |
ENSRNOT00000021171
|
Sfxn3
|
sideroflexin 3 |
| chr8_+_55279373 | 2.37 |
ENSRNOT00000064290
|
Ppp2r1b
|
protein phosphatase 2 scaffold subunit A beta |
| chr9_-_23352668 | 2.34 |
ENSRNOT00000075279
|
Mut
|
methylmalonyl CoA mutase |
| chr7_-_70498992 | 2.29 |
ENSRNOT00000067774
ENSRNOT00000079327 |
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase type 2 gamma |
| chr4_-_181477281 | 2.22 |
ENSRNOT00000055463
|
Mansc4
|
MANSC domain containing 4 |
| chr7_-_140417530 | 2.19 |
ENSRNOT00000077884
|
Fkbp11
|
FK506 binding protein 11 |
| chr3_+_75906945 | 2.18 |
ENSRNOT00000047110
|
Olr586
|
olfactory receptor 586 |
| chr19_+_46761570 | 2.16 |
ENSRNOT00000058779
|
Wwox
|
WW domain-containing oxidoreductase |
| chr3_-_57104030 | 2.14 |
ENSRNOT00000064203
|
Tlk1
|
tousled-like kinase 1 |
| chr15_+_39779648 | 2.14 |
ENSRNOT00000084505
|
Cab39l
|
calcium binding protein 39-like |
| chr4_-_115453659 | 2.14 |
ENSRNOT00000065847
|
Tex261
|
testis expressed 261 |
| chr2_-_165600748 | 2.10 |
ENSRNOT00000013216
|
Ift80
|
intraflagellar transport 80 |
| chrX_+_145357553 | 2.10 |
ENSRNOT00000033723
|
Magec2
|
MAGE family member C2 |
| chr4_+_45567573 | 2.05 |
ENSRNOT00000089824
|
Ankrd7
|
ankyrin repeat domain 7 |
| chr3_-_123206828 | 2.02 |
ENSRNOT00000030192
|
Ddrgk1
|
DDRGK domain containing 1 |
| chr10_+_3218466 | 2.02 |
ENSRNOT00000093629
ENSRNOT00000093338 ENSRNOT00000077695 |
Ntan1
|
N-terminal asparagine amidase |
| chr7_-_69982592 | 1.99 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr1_+_201117630 | 1.97 |
ENSRNOT00000077908
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr19_+_55880561 | 1.92 |
ENSRNOT00000047754
ENSRNOT00000091712 |
Spg7
|
SPG7, paraplegin matrix AAA peptidase subunit |
| chr13_-_47154292 | 1.90 |
ENSRNOT00000005284
|
Cd55
|
CD55 molecule, decay accelerating factor for complement |
| chr10_+_109533487 | 1.89 |
ENSRNOT00000054975
|
Fscn2
|
fascin actin-bundling protein 2, retinal |
| chr18_+_30435119 | 1.89 |
ENSRNOT00000027190
|
Pcdhb8
|
protocadherin beta 8 |
| chr17_-_45154355 | 1.89 |
ENSRNOT00000084976
|
Zkscan4
|
zinc finger with KRAB and SCAN domains 4 |
| chr1_-_276228574 | 1.89 |
ENSRNOT00000021746
|
Gucy2g
|
guanylate cyclase 2G |
| chr3_-_78066123 | 1.88 |
ENSRNOT00000073165
|
Olr690
|
olfactory receptor 690 |
| chr1_+_101152734 | 1.87 |
ENSRNOT00000028022
|
Pih1d1
|
PIH1 domain containing 1 |
| chr1_-_242440885 | 1.87 |
ENSRNOT00000076537
|
Pip5k1b
|
phosphatidylinositol-4-phosphate 5-kinase type 1 beta |
| chr1_+_168489077 | 1.85 |
ENSRNOT00000021191
|
Olr96
|
olfactory receptor 96 |
| chr4_+_113782206 | 1.84 |
ENSRNOT00000009701
|
M1ap
|
meiosis 1 associated protein |
| chr2_-_148722263 | 1.81 |
ENSRNOT00000017868
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr7_-_142063212 | 1.80 |
ENSRNOT00000089912
|
Slc11a2
|
solute carrier family 11 member 2 |
| chrX_+_112311251 | 1.79 |
ENSRNOT00000086698
|
AABR07040855.1
|
|
| chrX_-_135342996 | 1.79 |
ENSRNOT00000084848
ENSRNOT00000008503 |
Aifm1
|
apoptosis inducing factor, mitochondria associated 1 |
| chr4_-_51003117 | 1.76 |
ENSRNOT00000034936
|
Tas2r118
|
taste receptor, type 2, member 118 |
| chr3_-_91839009 | 1.75 |
ENSRNOT00000083703
|
Ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr5_+_155794229 | 1.75 |
ENSRNOT00000018127
|
LOC690206
|
hypothetical protein LOC690206 |
| chr1_-_242441247 | 1.75 |
ENSRNOT00000068645
|
Pip5k1b
|
phosphatidylinositol-4-phosphate 5-kinase type 1 beta |
| chr18_+_30387937 | 1.73 |
ENSRNOT00000027210
|
Pcdhb4
|
protocadherin beta 4 |
| chr1_+_69866069 | 1.72 |
ENSRNOT00000083609
|
LOC102547412
|
zinc finger protein 419-like |
| chr14_-_84751886 | 1.72 |
ENSRNOT00000078838
|
Mtmr3
|
myotubularin related protein 3 |
| chr12_+_12859661 | 1.67 |
ENSRNOT00000001404
|
Usp42
|
ubiquitin specific peptidase 42 |
| chr16_-_70998575 | 1.59 |
ENSRNOT00000019935
|
Kcnu1
|
potassium calcium-activated channel subfamily U member 1 |
| chr8_-_50277797 | 1.57 |
ENSRNOT00000082508
|
Pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr1_-_212021567 | 1.57 |
ENSRNOT00000029027
|
Cfap46
|
cilia and flagella associated protein 46 |
| chr12_+_2170630 | 1.56 |
ENSRNOT00000071928
|
Pet100
|
PET100 homolog |
| chr4_+_88048267 | 1.52 |
ENSRNOT00000044913
|
Vom1r81
|
vomeronasal 1 receptor 81 |
| chr1_-_91663467 | 1.52 |
ENSRNOT00000033396
|
Faap24
|
Fanconi anemia core complex associated protein 24 |
| chr5_+_159612762 | 1.52 |
ENSRNOT00000012147
|
Spata21
|
spermatogenesis associated 21 |
| chr1_+_101427195 | 1.49 |
ENSRNOT00000028271
|
Gys1
|
glycogen synthase 1 |
| chr5_+_155660553 | 1.46 |
ENSRNOT00000081893
|
Wnt4
|
wingless-type MMTV integration site family, member 4 |
| chr1_+_169115981 | 1.45 |
ENSRNOT00000067478
|
Olr135
|
olfactory receptor 135 |
| chr6_-_10592454 | 1.36 |
ENSRNOT00000020600
|
Pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr7_+_70335061 | 1.36 |
ENSRNOT00000072176
ENSRNOT00000084933 |
Cyp27b1
|
cytochrome P450, family 27, subfamily b, polypeptide 1 |
| chr1_-_263845762 | 1.35 |
ENSRNOT00000017268
|
Erlin1
|
ER lipid raft associated 1 |
| chr15_-_9086282 | 1.34 |
ENSRNOT00000008989
|
Thrb
|
thyroid hormone receptor beta |
| chr3_+_11587941 | 1.32 |
ENSRNOT00000071505
|
Dpm2
|
dolichyl-phosphate mannosyltransferase subunit 2, regulatory |
| chr13_-_99531959 | 1.32 |
ENSRNOT00000005059
|
Wdr26
|
WD repeat domain 26 |
| chr1_+_217039755 | 1.30 |
ENSRNOT00000091603
|
LOC102552318
|
actin-like |
| chr7_-_9465527 | 1.29 |
ENSRNOT00000044882
|
Olr1069
|
olfactory receptor 1069 |
| chr6_-_24201864 | 1.27 |
ENSRNOT00000080972
|
AABR07063248.1
|
|
| chr18_+_30880020 | 1.25 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr7_-_59139910 | 1.22 |
ENSRNOT00000032374
|
4933416C03Rik
|
RIKEN cDNA 4933416C03 gene |
| chr10_-_13892997 | 1.20 |
ENSRNOT00000004192
|
Traf7
|
TNF receptor associated factor 7 |
| chr4_-_146839397 | 1.19 |
ENSRNOT00000010338
|
Vgll4
|
vestigial-like family member 4 |
| chr4_+_87353254 | 1.15 |
ENSRNOT00000073670
|
Vom1r67
|
vomeronasal 1 receptor 67 |
| chr1_-_103426467 | 1.08 |
ENSRNOT00000045792
|
Mrgprb4
|
MAS-related GPR, member B4 |
| chr10_+_104582955 | 1.07 |
ENSRNOT00000009733
|
Unk
|
unkempt family zinc finger |
| chr3_+_76468294 | 1.04 |
ENSRNOT00000037779
|
Olr619
|
olfactory receptor 619 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfcp2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.6 | GO:0046223 | toxin catabolic process(GO:0009407) mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) secondary metabolite catabolic process(GO:0090487) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 3.9 | 11.8 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 3.8 | 15.0 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 3.3 | 22.8 | GO:0048143 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) astrocyte activation(GO:0048143) |
| 3.1 | 9.4 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 3.1 | 15.5 | GO:0036233 | glycine import(GO:0036233) |
| 2.8 | 11.3 | GO:0032919 | spermine acetylation(GO:0032919) |
| 2.8 | 8.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 2.8 | 16.6 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 2.6 | 7.9 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 2.6 | 10.3 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 2.4 | 7.1 | GO:0015692 | nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) |
| 1.9 | 13.6 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 1.9 | 5.6 | GO:0031439 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
| 1.7 | 11.8 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 1.6 | 6.4 | GO:0072249 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 1.4 | 7.0 | GO:0009758 | carbohydrate utilization(GO:0009758) |
| 1.3 | 4.0 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 1.3 | 13.2 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 1.3 | 3.9 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 1.3 | 5.2 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 1.3 | 16.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 1.3 | 7.6 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 1.2 | 3.6 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 1.1 | 6.7 | GO:0098928 | synaptic vesicle targeting(GO:0016080) presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 1.1 | 5.6 | GO:0035973 | aggrephagy(GO:0035973) |
| 1.1 | 20.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 1.0 | 6.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 1.0 | 4.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) oxygen metabolic process(GO:0072592) |
| 1.0 | 4.0 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 1.0 | 2.9 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 1.0 | 2.9 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.9 | 1.9 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.9 | 3.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.9 | 2.7 | GO:0042125 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.8 | 4.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.8 | 2.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.7 | 5.1 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.7 | 8.9 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.7 | 2.7 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.7 | 1.4 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) |
| 0.7 | 2.0 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.7 | 6.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.6 | 3.7 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.6 | 3.5 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.6 | 4.0 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.6 | 18.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.5 | 2.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.5 | 3.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.5 | 3.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.5 | 1.5 | GO:0060748 | female sex determination(GO:0030237) tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) positive regulation of dermatome development(GO:0061184) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.5 | 1.9 | GO:2000563 | regulation of complement activation, classical pathway(GO:0030450) positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.5 | 2.4 | GO:0006842 | tricarboxylic acid transport(GO:0006842) |
| 0.5 | 1.4 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.4 | 4.8 | GO:0071679 | cytoskeletal anchoring at plasma membrane(GO:0007016) commissural neuron axon guidance(GO:0071679) |
| 0.4 | 13.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.4 | 7.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.4 | 7.0 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.4 | 1.8 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.3 | 2.6 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 5.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.3 | 0.6 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.3 | 3.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 0.9 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.3 | 1.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.3 | 1.8 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.3 | 2.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 0.8 | GO:0010034 | response to acetate(GO:0010034) |
| 0.3 | 0.8 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.2 | 4.5 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.2 | 3.5 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.2 | 0.7 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.2 | 17.5 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.2 | 5.7 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.2 | 6.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 0.6 | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) dUMP metabolic process(GO:0046078) |
| 0.2 | 2.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 2.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 0.8 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 1.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 2.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.2 | 5.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 0.8 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 0.6 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.2 | 0.6 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.2 | 5.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 0.7 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.1 | 2.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 8.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 1.8 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 1.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 14.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 3.7 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 4.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 1.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.5 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 3.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 5.2 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 1.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 1.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 8.7 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 1.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 1.9 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 1.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 2.7 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.9 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 0.3 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 1.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 2.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 3.1 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 2.9 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 1.6 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 1.6 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.5 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.7 | GO:0070613 | regulation of protein processing(GO:0070613) |
| 0.0 | 3.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 5.4 | GO:0030522 | intracellular receptor signaling pathway(GO:0030522) |
| 0.0 | 0.8 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 2.2 | 6.7 | GO:0044317 | rod spherule(GO:0044317) |
| 2.1 | 8.4 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-2 complex(GO:0005607) laminin-10 complex(GO:0043259) |
| 1.5 | 15.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 1.4 | 5.6 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 1.3 | 6.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 1.1 | 14.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.7 | 3.7 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.7 | 4.8 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.7 | 11.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.6 | 1.9 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.6 | 3.5 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.6 | 4.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.5 | 3.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.5 | 9.3 | GO:0001741 | XY body(GO:0001741) |
| 0.5 | 7.4 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.5 | 2.7 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.4 | 5.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.4 | 1.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.4 | 2.6 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.4 | 5.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 3.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 8.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.4 | 2.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.3 | 8.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 1.9 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.2 | 3.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 2.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 17.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 0.8 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 9.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.6 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 0.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 3.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 7.6 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 11.8 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 5.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 4.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 13.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 5.2 | GO:1903293 | phosphatase complex(GO:1903293) |
| 0.1 | 0.6 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.0 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 2.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 3.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 5.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.6 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 16.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 9.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 0.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 4.0 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 5.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 5.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 3.5 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 24.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 5.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 36.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 3.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.1 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 4.3 | GO:0031966 | mitochondrial membrane(GO:0031966) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 3.1 | 9.4 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 3.0 | 8.9 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 2.7 | 29.9 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 2.6 | 5.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 2.3 | 7.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 2.3 | 7.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 2.3 | 11.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 2.0 | 8.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 2.0 | 16.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 2.0 | 7.9 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 1.9 | 5.6 | GO:0031249 | denatured protein binding(GO:0031249) |
| 1.4 | 5.6 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 1.3 | 11.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 1.2 | 3.6 | GO:0015203 | ornithine decarboxylase inhibitor activity(GO:0008073) polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 1.0 | 5.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 1.0 | 7.0 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.9 | 8.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.9 | 2.7 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 0.9 | 3.6 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.8 | 5.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.8 | 4.1 | GO:0016726 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.8 | 4.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.8 | 9.5 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.8 | 2.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.7 | 3.7 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.7 | 2.7 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.6 | 7.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.6 | 3.6 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.6 | 4.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.6 | 2.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.6 | 11.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.5 | 7.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.5 | 2.4 | GO:0015142 | tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.5 | 17.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.4 | 4.0 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.4 | 1.3 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.4 | 9.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.4 | 13.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.4 | 4.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.4 | 1.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.4 | 1.6 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 3.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.4 | 6.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 1.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 6.8 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.3 | 15.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 2.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 2.6 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 7.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.3 | 16.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.3 | 0.8 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 1.9 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.2 | 13.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 0.6 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 3.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 1.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 6.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 1.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.2 | 4.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 3.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 3.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 2.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 24.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.2 | 14.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 0.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 1.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 4.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 11.3 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 3.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 3.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 5.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 2.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 2.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 2.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 3.7 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 8.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 2.0 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 21.3 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 3.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 2.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 14.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.7 | 4.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.6 | 11.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.5 | 5.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 12.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 3.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 21.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 9.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 11.2 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.2 | 13.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.2 | 3.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 2.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 7.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 5.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 3.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 0.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 2.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 17.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 3.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 5.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.4 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 13.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 1.0 | 36.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.9 | 22.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.8 | 9.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.6 | 17.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 3.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.5 | 10.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.5 | 7.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 11.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.4 | 15.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 6.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.3 | 11.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 16.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.2 | 3.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 3.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 2.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 6.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 2.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 7.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 5.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 11.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 5.6 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 2.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 10.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 4.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 3.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 3.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 6.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 2.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.5 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.9 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.9 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |