Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
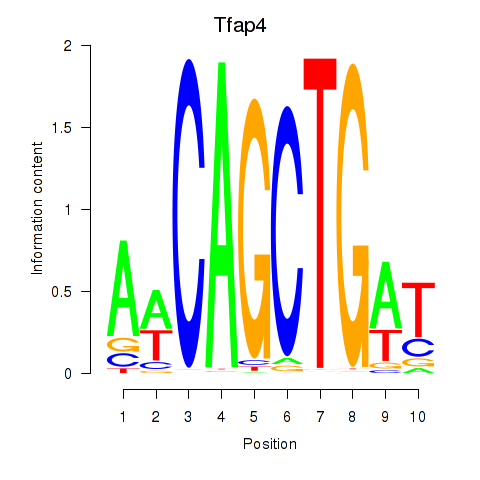
Results for Tfap4
Z-value: 1.02
Transcription factors associated with Tfap4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap4
|
ENSRNOG00000005227 | transcription factor AP-4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap4 | rn6_v1_chr10_+_11206226_11206226 | -0.30 | 7.2e-08 | Click! |
Activity profile of Tfap4 motif
Sorted Z-values of Tfap4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_55178289 | 43.66 |
ENSRNOT00000059127
|
Cryab
|
crystallin, alpha B |
| chr6_-_26624092 | 42.51 |
ENSRNOT00000008113
|
Trim54
|
tripartite motif-containing 54 |
| chr4_+_144382945 | 38.87 |
ENSRNOT00000007601
|
Cav3
|
caveolin 3 |
| chr17_+_32973695 | 38.59 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr13_-_84331905 | 35.76 |
ENSRNOT00000004965
|
Dusp27
|
dual specificity phosphatase 27 (putative) |
| chr18_+_79406381 | 34.09 |
ENSRNOT00000022303
ENSRNOT00000058295 ENSRNOT00000058296 ENSRNOT00000022280 |
Mbp
|
myelin basic protein |
| chr1_-_89560469 | 30.82 |
ENSRNOT00000079091
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr7_-_83670356 | 30.56 |
ENSRNOT00000005584
|
Sybu
|
syntabulin |
| chr4_-_89151184 | 30.10 |
ENSRNOT00000010263
|
Nap1l5
|
nucleosome assembly protein 1-like 5 |
| chr14_+_37113210 | 29.91 |
ENSRNOT00000089094
|
Sgcb
|
sarcoglycan, beta |
| chrX_+_159158194 | 26.78 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr10_+_92628356 | 26.59 |
ENSRNOT00000072480
|
Myl4
|
myosin, light chain 4 |
| chr17_+_12762752 | 26.33 |
ENSRNOT00000044842
|
Diras2
|
DIRAS family GTPase 2 |
| chr7_-_141185710 | 25.79 |
ENSRNOT00000085033
|
Faim2
|
Fas apoptotic inhibitory molecule 2 |
| chr7_-_139063752 | 25.72 |
ENSRNOT00000072309
|
LOC102551901
|
protein lifeguard 2-like |
| chr2_+_114386019 | 25.66 |
ENSRNOT00000082148
|
AABR07009834.1
|
|
| chr8_-_55177818 | 24.94 |
ENSRNOT00000013960
|
Hspb2
|
heat shock protein family B (small) member 2 |
| chr6_-_105097054 | 24.76 |
ENSRNOT00000048606
|
Slc8a3
|
solute carrier family 8 member A3 |
| chr4_+_199916 | 24.74 |
ENSRNOT00000009317
|
Htr5a
|
5-hydroxytryptamine receptor 5A |
| chr3_+_168345152 | 23.72 |
ENSRNOT00000017654
|
Dok5
|
docking protein 5 |
| chr6_+_58467254 | 23.44 |
ENSRNOT00000065396
|
Etv1
|
ets variant 1 |
| chr16_-_40025401 | 21.89 |
ENSRNOT00000066639
|
Asb5
|
ankyrin repeat and SOCS box-containing 5 |
| chr9_+_81821346 | 21.20 |
ENSRNOT00000022234
|
Plcd4
|
phospholipase C, delta 4 |
| chr2_-_184289126 | 21.00 |
ENSRNOT00000081678
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chrX_+_92131209 | 20.78 |
ENSRNOT00000004462
|
Pabpc5
|
poly A binding protein, cytoplasmic 5 |
| chr3_-_61494778 | 20.64 |
ENSRNOT00000068018
|
Lnpk
|
lunapark, ER junction formation factor |
| chr10_+_99437436 | 20.31 |
ENSRNOT00000006254
|
Kcnj2
|
potassium voltage-gated channel subfamily J member 2 |
| chr2_-_210943620 | 19.19 |
ENSRNOT00000026750
|
Gpr61
|
G protein-coupled receptor 61 |
| chr7_+_70807867 | 19.10 |
ENSRNOT00000010639
|
Stac3
|
SH3 and cysteine rich domain 3 |
| chr10_-_95934345 | 18.72 |
ENSRNOT00000004349
|
Cacng1
|
calcium voltage-gated channel auxiliary subunit gamma 1 |
| chr4_-_482645 | 18.56 |
ENSRNOT00000062073
ENSRNOT00000071713 |
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr20_+_20236151 | 18.43 |
ENSRNOT00000079630
|
Ank3
|
ankyrin 3 |
| chr18_-_5314511 | 17.98 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr5_+_151692108 | 17.55 |
ENSRNOT00000086144
|
Fam46b
|
family with sequence similarity 46, member B |
| chr8_-_87419564 | 17.43 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chrX_-_134866210 | 17.28 |
ENSRNOT00000005331
|
Apln
|
apelin |
| chr15_+_19547871 | 17.26 |
ENSRNOT00000036235
|
Gpr137c
|
G protein-coupled receptor 137C |
| chr17_-_79085076 | 16.66 |
ENSRNOT00000057851
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr17_+_1679627 | 16.59 |
ENSRNOT00000025801
|
Habp4
|
hyaluronan binding protein 4 |
| chr8_-_43336304 | 16.24 |
ENSRNOT00000036054
|
RGD1311744
|
similar to RIKEN cDNA 5830475I06 |
| chr15_+_110114148 | 15.65 |
ENSRNOT00000006264
|
Itgbl1
|
integrin subunit beta like 1 |
| chr1_+_171793782 | 15.39 |
ENSRNOT00000081351
|
Olfml1
|
olfactomedin-like 1 |
| chr3_+_150910398 | 15.32 |
ENSRNOT00000055310
|
Tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr5_+_164808323 | 14.85 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr15_+_43905099 | 14.73 |
ENSRNOT00000016568
|
Ebf2
|
early B-cell factor 2 |
| chr16_+_6962722 | 14.01 |
ENSRNOT00000023330
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr5_-_58484900 | 13.58 |
ENSRNOT00000012386
|
Fam214b
|
family with sequence similarity 214, member B |
| chr3_+_177310753 | 13.45 |
ENSRNOT00000031448
|
Myt1
|
myelin transcription factor 1 |
| chr17_+_77167740 | 13.23 |
ENSRNOT00000042881
|
Optn
|
optineurin |
| chr6_-_102047758 | 13.13 |
ENSRNOT00000012101
|
Atp6v1d
|
ATPase H+ transporting V1 subunit D |
| chr3_-_81304181 | 12.83 |
ENSRNOT00000079746
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr18_-_14016713 | 12.80 |
ENSRNOT00000041125
|
Nol4
|
nucleolar protein 4 |
| chr8_+_130350510 | 12.58 |
ENSRNOT00000073376
|
Ss18l2
|
SS18 like 2 |
| chr4_-_50860756 | 12.37 |
ENSRNOT00000068404
|
Cadps2
|
calcium dependent secretion activator 2 |
| chr9_-_32019205 | 12.34 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr1_+_201055644 | 12.16 |
ENSRNOT00000054937
ENSRNOT00000047161 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr1_-_215846911 | 12.00 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr18_-_56870801 | 11.78 |
ENSRNOT00000087188
ENSRNOT00000032189 |
Arhgef37
|
Rho guanine nucleotide exchange factor 37 |
| chr9_+_111028575 | 11.49 |
ENSRNOT00000043451
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_-_148932878 | 11.33 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chrX_+_71342775 | 10.77 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr7_+_73588163 | 10.66 |
ENSRNOT00000015707
|
Kcns2
|
potassium voltage-gated channel, modifier subfamily S, member 2 |
| chr9_+_111028824 | 10.66 |
ENSRNOT00000041418
ENSRNOT00000056457 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr8_+_49676540 | 10.62 |
ENSRNOT00000022032
ENSRNOT00000082205 |
Fxyd6
|
FXYD domain-containing ion transport regulator 6 |
| chr1_+_77994203 | 10.39 |
ENSRNOT00000002044
|
Napa
|
NSF attachment protein alpha |
| chr10_-_64642292 | 10.07 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chr3_-_64024205 | 10.02 |
ENSRNOT00000037015
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr7_+_140383397 | 9.88 |
ENSRNOT00000090760
|
Ccdc65
|
coiled-coil domain containing 65 |
| chrX_+_63520991 | 9.75 |
ENSRNOT00000071590
|
Apoo
|
apolipoprotein O |
| chr2_-_98610368 | 9.54 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr13_+_52662996 | 9.39 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr8_+_45797315 | 8.77 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chr11_-_24641820 | 8.75 |
ENSRNOT00000044081
ENSRNOT00000048854 |
App
|
amyloid beta precursor protein |
| chr8_-_127900463 | 8.69 |
ENSRNOT00000078303
|
Slc22a13
|
solute carrier family 22 member 13 |
| chr4_+_114876770 | 8.63 |
ENSRNOT00000068102
ENSRNOT00000078175 |
Dctn1
|
dynactin subunit 1 |
| chr2_+_189423559 | 8.54 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
| chr4_-_157358262 | 8.42 |
ENSRNOT00000021481
|
Gnb3
|
G protein subunit beta 3 |
| chr7_+_64672722 | 8.15 |
ENSRNOT00000064448
ENSRNOT00000005539 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr2_-_140618405 | 7.94 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr20_+_3556975 | 7.93 |
ENSRNOT00000089417
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr12_-_25638797 | 7.67 |
ENSRNOT00000002033
|
Gatsl2
|
GATS protein-like 2 |
| chr14_+_42007312 | 7.37 |
ENSRNOT00000063985
|
Atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr1_+_141218095 | 7.32 |
ENSRNOT00000051411
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr5_-_173318313 | 7.27 |
ENSRNOT00000039117
|
Cptp
|
ceramide-1-phosphate transfer protein |
| chr6_-_21135880 | 7.14 |
ENSRNOT00000051239
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chr1_-_277768368 | 7.09 |
ENSRNOT00000023113
|
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr7_-_144322240 | 6.92 |
ENSRNOT00000089290
|
LOC100912282
|
calcium-binding and coiled-coil domain-containing protein 1-like |
| chr2_-_227145185 | 6.88 |
ENSRNOT00000083189
|
AABR07013202.1
|
|
| chr3_-_72769912 | 6.86 |
ENSRNOT00000043742
|
Olr439
|
olfactory receptor 439 |
| chr17_-_55346279 | 6.83 |
ENSRNOT00000025037
|
Svil
|
supervillin |
| chr1_-_215553451 | 6.57 |
ENSRNOT00000027407
|
Ctsd
|
cathepsin D |
| chr15_-_61772516 | 6.55 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chr8_-_127912860 | 6.51 |
ENSRNOT00000040498
|
LOC685081
|
similar to solute carrier family 22 (organic cation transporter), member 13 |
| chr11_-_84047497 | 6.29 |
ENSRNOT00000088821
ENSRNOT00000058092 |
Ap2m1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr10_-_49007089 | 6.20 |
ENSRNOT00000073706
|
Mapk7
|
mitogen-activated protein kinase 7 |
| chr7_+_120153184 | 6.13 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr7_-_44121130 | 6.11 |
ENSRNOT00000005706
|
Nts
|
neurotensin |
| chr19_-_33424955 | 6.09 |
ENSRNOT00000016558
ENSRNOT00000089569 |
Ttc29
|
tetratricopeptide repeat domain 29 |
| chr4_-_157358007 | 5.95 |
ENSRNOT00000090898
|
Gnb3
|
G protein subunit beta 3 |
| chr7_-_145450233 | 5.56 |
ENSRNOT00000092974
ENSRNOT00000021523 |
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr14_-_37871051 | 5.49 |
ENSRNOT00000003087
|
Slain2
|
SLAIN motif family, member 2 |
| chr2_-_53827175 | 5.49 |
ENSRNOT00000078158
|
RGD1305938
|
similar to expressed sequence AW549877 |
| chr12_+_40553741 | 5.44 |
ENSRNOT00000057396
|
LOC100360238
|
rCG21419-like |
| chr13_-_53224768 | 5.41 |
ENSRNOT00000091015
|
Camsap2
|
calmodulin regulated spectrin-associated protein family, member 2 |
| chr19_-_41349681 | 5.40 |
ENSRNOT00000080694
ENSRNOT00000059147 |
Hydin
|
Hydin, axonemal central pair apparatus protein |
| chr4_+_181243338 | 5.39 |
ENSRNOT00000034051
|
Smco2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr7_+_77678968 | 5.39 |
ENSRNOT00000006917
|
Atp6v1c1
|
ATPase H+ transporting V1 subunit C1 |
| chr1_+_252906234 | 5.38 |
ENSRNOT00000031025
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr9_+_17705212 | 5.38 |
ENSRNOT00000026728
|
Tmem63b
|
transmembrane protein 63B |
| chr1_-_281756159 | 5.38 |
ENSRNOT00000013170
|
Prlhr
|
prolactin releasing hormone receptor |
| chr4_-_129544429 | 5.35 |
ENSRNOT00000091507
ENSRNOT00000083461 ENSRNOT00000087215 |
Tmf1
|
TATA element modulatory factor 1 |
| chr2_-_244370983 | 5.24 |
ENSRNOT00000021458
|
Rap1gds1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr6_-_22138286 | 5.20 |
ENSRNOT00000007607
|
Yipf4
|
Yip1 domain family, member 4 |
| chr9_-_85528860 | 5.14 |
ENSRNOT00000063834
ENSRNOT00000088963 |
Wdfy1
|
WD repeat and FYVE domain containing 1 |
| chr17_-_10208360 | 5.03 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr10_+_14828597 | 4.88 |
ENSRNOT00000025434
|
Tekt4
|
tektin 4 |
| chr10_-_89338739 | 4.63 |
ENSRNOT00000073923
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chrX_+_62754634 | 4.56 |
ENSRNOT00000016669
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr10_+_55642070 | 4.37 |
ENSRNOT00000008507
|
Borcs6
|
BLOC-1 related complex subunit 6 |
| chr14_-_114649173 | 4.27 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr5_+_172364421 | 4.24 |
ENSRNOT00000018769
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr6_+_137164535 | 4.13 |
ENSRNOT00000018225
|
Inf2
|
inverted formin, FH2 and WH2 domain containing |
| chr15_-_4022223 | 4.10 |
ENSRNOT00000081997
|
Zswim8
|
zinc finger, SWIM-type containing 8 |
| chr10_+_85301875 | 4.05 |
ENSRNOT00000080935
|
Socs7
|
suppressor of cytokine signaling 7 |
| chr1_-_214844858 | 4.03 |
ENSRNOT00000046344
|
Tollip
|
toll interacting protein |
| chr8_-_48582353 | 3.83 |
ENSRNOT00000011582
|
Pdzd3
|
PDZ domain containing 3 |
| chr18_+_61490031 | 3.64 |
ENSRNOT00000022958
|
Sec11c
|
SEC11 homolog C, signal peptidase complex subunit |
| chr14_-_8548310 | 3.48 |
ENSRNOT00000092436
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr19_+_24329023 | 3.48 |
ENSRNOT00000065558
|
Tbc1d9
|
TBC1 domain family member 9 |
| chr3_-_46153371 | 3.43 |
ENSRNOT00000085885
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr16_-_62294769 | 3.34 |
ENSRNOT00000020663
|
Ppp2cb
|
protein phosphatase 2 catalytic subunit beta |
| chr16_-_32540217 | 3.27 |
ENSRNOT00000015130
|
Hpf1
|
histone PARylation factor 1 |
| chr11_-_28672956 | 3.19 |
ENSRNOT00000002137
|
LOC100363136
|
hypothetical protein LOC100363136 |
| chr8_-_61836796 | 3.03 |
ENSRNOT00000064903
ENSRNOT00000084009 |
Commd4
|
COMM domain containing 4 |
| chr1_-_89509343 | 2.85 |
ENSRNOT00000028637
|
Fxyd3
|
FXYD domain-containing ion transport regulator 3 |
| chr1_-_273758442 | 2.63 |
ENSRNOT00000088016
|
Xpnpep1
|
X-prolyl aminopeptidase 1 |
| chr10_-_65780349 | 2.53 |
ENSRNOT00000013220
|
Tmem199
|
transmembrane protein 199 |
| chr4_-_165810121 | 2.50 |
ENSRNOT00000048210
|
Tas2r104
|
taste receptor, type 2, member 104 |
| chrX_+_23668363 | 2.43 |
ENSRNOT00000004758
|
Gpr143
|
G protein-coupled receptor 143 |
| chr2_+_243840134 | 2.29 |
ENSRNOT00000093355
|
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr3_-_72740519 | 2.28 |
ENSRNOT00000040012
|
Olr436
|
olfactory receptor 436 |
| chr1_-_214971929 | 2.23 |
ENSRNOT00000027195
|
Mob2
|
MOB kinase activator 2 |
| chr15_-_5894854 | 2.21 |
ENSRNOT00000024427
|
Cd99l2
|
CD99 molecule-like 2 |
| chr8_-_71050853 | 2.09 |
ENSRNOT00000073571
|
Ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr9_-_120101098 | 2.03 |
ENSRNOT00000002917
|
Fam114a1l1
|
family with sequence similarity 114, member A1-like 1 |
| chr3_+_111097326 | 1.97 |
ENSRNOT00000018717
|
Vps18
|
VPS18 CORVET/HOPS core subunit |
| chr15_+_5265307 | 1.92 |
ENSRNOT00000031807
|
LOC108348081
|
CD99 antigen-like protein 2 |
| chr4_+_159483131 | 1.92 |
ENSRNOT00000089513
|
RGD1311164
|
similar to DNA segment, Chr 6, Wayne State University 163, expressed |
| chr5_-_101166651 | 1.69 |
ENSRNOT00000078862
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr8_+_70884491 | 1.68 |
ENSRNOT00000072939
|
Ubap1l
|
ubiquitin associated protein 1-like |
| chr3_-_5617225 | 1.68 |
ENSRNOT00000008327
|
Tmem8c
|
transmembrane protein 8C |
| chr19_+_52032886 | 1.61 |
ENSRNOT00000019923
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr8_-_55491152 | 1.60 |
ENSRNOT00000014965
|
LOC100359479
|
rCG58364-like |
| chr14_-_82287706 | 1.59 |
ENSRNOT00000080695
|
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr1_-_273758247 | 1.51 |
ENSRNOT00000033148
|
Xpnpep1
|
X-prolyl aminopeptidase 1 |
| chr16_+_32457521 | 1.37 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr2_-_211235440 | 1.34 |
ENSRNOT00000033015
ENSRNOT00000091896 |
Sars
|
seryl-tRNA synthetase |
| chr12_+_2054680 | 1.24 |
ENSRNOT00000001290
|
Mcoln1
|
mucolipin 1 |
| chr7_+_11838639 | 1.12 |
ENSRNOT00000025878
|
Ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr3_-_60460724 | 1.12 |
ENSRNOT00000024706
|
Chrna1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr11_-_36533073 | 1.11 |
ENSRNOT00000033486
|
Lca5l
|
LCA5L, lebercilin like |
| chr19_+_14392423 | 1.08 |
ENSRNOT00000018880
|
Tom1
|
target of myb1 membrane trafficking protein |
| chr20_-_11626876 | 1.03 |
ENSRNOT00000001635
|
LOC100361664
|
keratin associated protein 12-1-like |
| chr10_+_65780494 | 0.83 |
ENSRNOT00000013100
|
Poldip2
|
DNA polymerase delta interacting protein 2 |
| chr13_-_74740458 | 0.78 |
ENSRNOT00000006548
|
Tex35
|
testis expressed 35 |
| chr7_-_125497691 | 0.74 |
ENSRNOT00000049445
|
AABR07058578.1
|
|
| chr2_+_198683159 | 0.73 |
ENSRNOT00000028793
|
Txnip
|
thioredoxin interacting protein |
| chr8_-_22874637 | 0.71 |
ENSRNOT00000064551
ENSRNOT00000090424 |
Dock6
|
dedicator of cytokinesis 6 |
| chr1_+_282188110 | 0.67 |
ENSRNOT00000013845
|
Fam45a
|
family with sequence similarity 45, member A |
| chr12_-_16934706 | 0.50 |
ENSRNOT00000001720
|
Mafk
|
MAF bZIP transcription factor K |
| chr8_+_41987849 | 0.47 |
ENSRNOT00000073800
|
LOC100911999
|
olfactory receptor 143-like |
| chr3_-_72895740 | 0.43 |
ENSRNOT00000012568
|
Fads2l1
|
fatty acid desaturase 2-like 1 |
| chr19_-_37525762 | 0.43 |
ENSRNOT00000023606
|
Atp6v0d1
|
ATPase H+ transporting V0 subunit D1 |
| chr3_+_16001207 | 0.43 |
ENSRNOT00000041987
|
Olr402
|
olfactory receptor 402 |
| chr17_-_29360822 | 0.40 |
ENSRNOT00000080031
|
Fars2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr10_+_60974225 | 0.40 |
ENSRNOT00000042349
|
Olr1511
|
olfactory receptor 1511 |
| chr5_-_33892462 | 0.36 |
ENSRNOT00000009334
|
Atp6v0d2
|
ATPase H+ transporting V0 subunit D2 |
| chrX_-_26376467 | 0.33 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr11_-_17340373 | 0.22 |
ENSRNOT00000089762
|
AABR07033324.1
|
|
| chr7_+_13039781 | 0.20 |
ENSRNOT00000067771
ENSRNOT00000090048 |
Mier2
|
mesoderm induction early response 1, family member 2 |
| chr13_-_73704678 | 0.17 |
ENSRNOT00000005280
|
Tor1aip1
|
torsin 1A interacting protein 1 |
| chrX_-_119532965 | 0.09 |
ENSRNOT00000073677
|
AABR07041109.1
|
|
| chr5_+_156603725 | 0.06 |
ENSRNOT00000075852
|
Hp1bp3
|
heterochromatin protein 1, binding protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.4 | 34.1 | GO:1904685 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) positive regulation of metalloendopeptidase activity(GO:1904685) |
| 9.7 | 38.9 | GO:0035995 | detection of muscle stretch(GO:0035995) positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 7.0 | 21.0 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 6.2 | 24.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 6.2 | 30.8 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 5.5 | 22.1 | GO:0031179 | peptide modification(GO:0031179) |
| 5.5 | 43.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 5.0 | 14.9 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 4.6 | 18.4 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 4.3 | 17.3 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 4.2 | 4.2 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 4.1 | 24.7 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 4.1 | 45.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 3.5 | 14.0 | GO:1902730 | positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 3.4 | 10.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 3.1 | 9.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 3.0 | 29.9 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 2.9 | 20.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 2.6 | 10.4 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 2.5 | 7.4 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 2.1 | 10.6 | GO:0061743 | motor learning(GO:0061743) |
| 1.8 | 5.4 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 1.8 | 8.8 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 1.7 | 8.6 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 1.7 | 12.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.7 | 13.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.6 | 19.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 1.5 | 12.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 1.5 | 16.2 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 1.5 | 7.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 1.4 | 23.7 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 1.4 | 25.8 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 1.3 | 5.4 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 1.3 | 3.8 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) |
| 1.2 | 6.2 | GO:0071499 | cellular response to laminar fluid shear stress(GO:0071499) |
| 1.2 | 6.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 1.2 | 13.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 1.2 | 6.1 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 1.2 | 12.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.1 | 14.7 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 1.1 | 12.2 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 1.1 | 3.3 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 1.1 | 15.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 1.1 | 42.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 1.0 | 4.9 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.9 | 6.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.8 | 5.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.8 | 20.0 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.8 | 7.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.8 | 10.7 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.7 | 5.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.7 | 7.9 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.7 | 18.7 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.7 | 26.8 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.7 | 5.2 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.6 | 9.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.6 | 2.4 | GO:1903056 | regulation of melanosome organization(GO:1903056) |
| 0.6 | 20.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.6 | 4.6 | GO:1904116 | response to vasopressin(GO:1904116) |
| 0.6 | 1.1 | GO:0070142 | synaptic vesicle budding(GO:0070142) |
| 0.6 | 1.7 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.5 | 1.6 | GO:0034034 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.5 | 4.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.5 | 21.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.5 | 4.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.4 | 4.3 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.4 | 5.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.4 | 13.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.4 | 10.0 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.3 | 3.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.3 | 1.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.3 | 3.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.3 | 13.1 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.3 | 1.6 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.3 | 6.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.3 | 22.7 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.3 | 2.0 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.3 | 21.9 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.3 | 28.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 4.0 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) protein localization to endosome(GO:0036010) |
| 0.2 | 18.0 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.2 | 6.2 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.2 | 7.1 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.2 | 16.6 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.2 | 4.1 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.2 | 5.4 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 26.3 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.1 | 1.7 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.7 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 6.3 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 3.3 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 1.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.8 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 8.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.6 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.7 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 8.2 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.3 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 1.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 2.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 4.5 | GO:0006470 | protein dephosphorylation(GO:0006470) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 53.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 5.7 | 34.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 5.0 | 29.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 4.3 | 12.8 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 4.2 | 21.0 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 3.1 | 30.6 | GO:0097433 | dense body(GO:0097433) |
| 3.0 | 38.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 2.2 | 8.8 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 2.0 | 49.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 1.7 | 10.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.4 | 9.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.2 | 8.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.1 | 8.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 1.1 | 5.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 1.1 | 5.4 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.8 | 12.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.8 | 5.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.6 | 6.8 | GO:0043034 | costamere(GO:0043034) |
| 0.6 | 3.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.6 | 30.9 | GO:0031672 | A band(GO:0031672) |
| 0.6 | 13.9 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.5 | 14.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.5 | 20.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.4 | 7.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 44.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.4 | 6.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.4 | 18.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 24.8 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.4 | 16.6 | GO:0030017 | sarcomere(GO:0030017) |
| 0.4 | 1.1 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.3 | 2.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.3 | 2.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 7.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 22.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.3 | 7.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.2 | 24.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 4.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 2.0 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 21.5 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 44.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 1.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 5.5 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.1 | 5.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 10.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 3.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.5 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 1.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 6.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 13.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 7.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 7.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 7.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.4 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 4.8 | GO:0005813 | centrosome(GO:0005813) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 30.8 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 5.5 | 22.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 4.9 | 38.9 | GO:0071253 | connexin binding(GO:0071253) |
| 4.2 | 21.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 3.5 | 24.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 3.5 | 10.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 3.1 | 68.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 2.8 | 34.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 2.6 | 13.2 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 2.3 | 9.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 2.2 | 8.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 1.8 | 12.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 1.8 | 7.3 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 1.8 | 24.7 | GO:0051378 | serotonin binding(GO:0051378) |
| 1.6 | 20.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 1.2 | 1.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 1.1 | 7.9 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.1 | 15.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 1.0 | 6.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 1.0 | 5.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.9 | 20.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.9 | 4.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.9 | 30.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.8 | 32.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.7 | 12.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.6 | 5.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.6 | 30.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.6 | 31.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.6 | 5.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.5 | 8.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.5 | 3.8 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.5 | 1.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.4 | 5.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.4 | 7.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.4 | 18.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.3 | 6.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 10.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 7.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 13.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 6.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.3 | 8.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.3 | 8.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 4.0 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.3 | 6.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 2.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 6.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 26.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 1.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 8.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 4.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 7.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 2.4 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.1 | 0.5 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 13.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 13.1 | GO:0043492 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.1 | 25.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 9.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 4.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 3.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 12.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.3 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 2.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 6.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 6.4 | GO:0005549 | odorant binding(GO:0005549) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 32.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.9 | 23.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.6 | 25.8 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.6 | 23.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.6 | 12.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.6 | 28.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.5 | 14.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 6.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 6.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 8.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 12.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 4.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 8.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 8.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 7.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 6.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 5.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 6.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 24.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.5 | 53.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.2 | 34.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 1.1 | 44.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.0 | 24.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.8 | 9.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.7 | 19.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.7 | 6.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.7 | 9.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.7 | 21.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.6 | 12.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 14.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.4 | 5.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 1.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 10.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 3.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 6.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.2 | 7.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 8.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 4.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 2.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 5.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 6.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 11.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 5.9 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 1.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 3.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |