Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
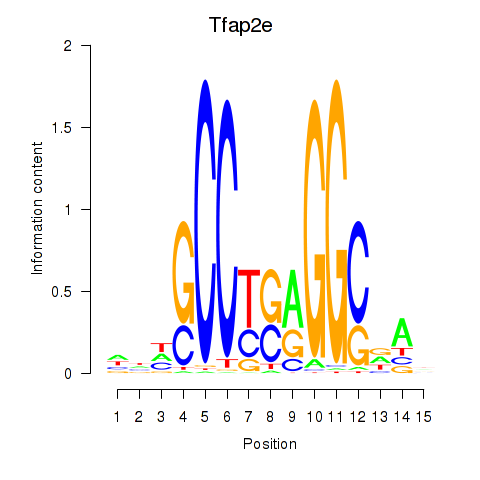
Results for Tfap2e
Z-value: 0.49
Transcription factors associated with Tfap2e
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2e
|
ENSRNOG00000027595 | transcription factor AP-2 epsilon |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2e | rn6_v1_chr5_-_144761787_144761787 | -0.12 | 3.0e-02 | Click! |
Activity profile of Tfap2e motif
Sorted Z-values of Tfap2e motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_100530183 | 26.27 |
ENSRNOT00000067754
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr10_+_86340940 | 24.78 |
ENSRNOT00000073486
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr3_+_5709236 | 18.27 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr10_-_98018014 | 11.84 |
ENSRNOT00000005367
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr18_-_55859333 | 10.11 |
ENSRNOT00000025989
|
Myoz3
|
myozenin 3 |
| chr9_+_99998275 | 8.62 |
ENSRNOT00000074395
|
Gpc1
|
glypican 1 |
| chr18_-_40716686 | 8.60 |
ENSRNOT00000000172
|
Cdo1
|
cysteine dioxygenase type 1 |
| chr4_-_113610243 | 6.14 |
ENSRNOT00000008813
|
Hk2
|
hexokinase 2 |
| chr7_-_77162148 | 5.93 |
ENSRNOT00000008350
|
Klf10
|
Kruppel-like factor 10 |
| chr8_-_50531423 | 4.87 |
ENSRNOT00000090985
ENSRNOT00000074942 |
Apoc3
|
apolipoprotein C3 |
| chrX_-_157095274 | 4.78 |
ENSRNOT00000084088
|
Abcd1
|
ATP binding cassette subfamily D member 1 |
| chr17_-_35037076 | 4.13 |
ENSRNOT00000090946
|
Dusp22
|
dual specificity phosphatase 22 |
| chr1_+_78933372 | 4.07 |
ENSRNOT00000029810
|
Ccdc8
|
coiled-coil domain containing 8 |
| chr17_+_11953552 | 3.85 |
ENSRNOT00000090782
|
Ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr4_+_120133713 | 3.57 |
ENSRNOT00000017240
|
Gata2
|
GATA binding protein 2 |
| chr10_-_85684138 | 3.08 |
ENSRNOT00000017989
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase type 2 beta |
| chr4_-_64831233 | 2.96 |
ENSRNOT00000079285
|
Dgki
|
diacylglycerol kinase, iota |
| chr7_-_140454268 | 2.72 |
ENSRNOT00000081468
|
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr8_+_53678777 | 2.71 |
ENSRNOT00000045944
|
Drd2
|
dopamine receptor D2 |
| chr10_+_83231409 | 2.54 |
ENSRNOT00000006230
|
Spop
|
speckle type BTB/POZ protein |
| chr17_-_10208360 | 2.33 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr10_+_105156632 | 2.29 |
ENSRNOT00000086288
|
Galr2
|
galanin receptor 2 |
| chr10_+_105156822 | 1.81 |
ENSRNOT00000012523
|
Galr2
|
galanin receptor 2 |
| chr8_+_22625874 | 1.78 |
ENSRNOT00000012269
|
Timm29
|
translocase of inner mitochondrial membrane 29 |
| chr20_-_3793985 | 1.57 |
ENSRNOT00000049540
ENSRNOT00000086293 |
RT1-CE16
|
RT1 class I, locus CE16 |
| chr1_+_173275336 | 1.51 |
ENSRNOT00000086302
|
AABR07004992.1
|
|
| chr1_+_79911516 | 1.46 |
ENSRNOT00000019075
|
Irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr2_+_189169877 | 1.21 |
ENSRNOT00000028225
|
She
|
Src homology 2 domain containing E |
| chr19_-_11326139 | 1.12 |
ENSRNOT00000025669
|
Mt3
|
metallothionein 3 |
| chr10_+_84031955 | 1.12 |
ENSRNOT00000009934
|
Hoxb13
|
homeo box B13 |
| chr10_+_86582612 | 0.97 |
ENSRNOT00000048349
|
Gsdma
|
gasdermin A |
| chr1_+_87045796 | 0.92 |
ENSRNOT00000027620
|
Lgals7
|
galectin 7 |
| chr14_+_81819415 | 0.90 |
ENSRNOT00000018515
ENSRNOT00000075863 ENSRNOT00000077060 |
Mxd4
|
Max dimerization protein 4 |
| chr9_-_115382445 | 0.81 |
ENSRNOT00000091316
|
Ptprm
|
protein tyrosine phosphatase, receptor type, M |
| chrX_+_21696772 | 0.73 |
ENSRNOT00000043559
|
Hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr9_-_24451435 | 0.69 |
ENSRNOT00000060805
|
Defb49
|
defensin beta 49 |
| chrX_-_73778595 | 0.52 |
ENSRNOT00000076081
ENSRNOT00000075926 ENSRNOT00000003782 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr1_-_168471161 | 0.37 |
ENSRNOT00000055227
|
Olr94
|
olfactory receptor 94 |
| chr8_+_108958046 | 0.28 |
ENSRNOT00000079618
ENSRNOT00000066917 |
Stag1
|
stromal antigen 1 |
| chr20_-_1984737 | 0.19 |
ENSRNOT00000040232
ENSRNOT00000051634 ENSRNOT00000079445 |
Gabbr1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr19_-_25801526 | 0.17 |
ENSRNOT00000003884
|
Nacc1
|
nucleus accumbens associated 1 |
| chr8_-_22625959 | 0.10 |
ENSRNOT00000012138
|
Yipf2
|
Yip1 domain family, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2e
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.3 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 4.1 | 24.8 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 2.9 | 8.6 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 2.9 | 8.6 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 2.6 | 26.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 2.4 | 11.8 | GO:0044691 | tooth eruption(GO:0044691) |
| 1.6 | 4.9 | GO:0010982 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 1.5 | 6.1 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 1.2 | 3.6 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 1.0 | 4.1 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 1.0 | 3.8 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.8 | 4.8 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.7 | 2.7 | GO:0042640 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) anagen(GO:0042640) regulation of anagen(GO:0051884) |
| 0.7 | 2.7 | GO:1900166 | glial cell-derived neurotrophic factor secretion(GO:0044467) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) negative regulation of dopamine receptor signaling pathway(GO:0060160) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.4 | 2.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 1.1 | GO:2000295 | cadmium ion homeostasis(GO:0055073) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.4 | 3.0 | GO:0046959 | habituation(GO:0046959) |
| 0.3 | 2.5 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.2 | 4.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.2 | 1.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 5.9 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.2 | 0.5 | GO:0060816 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 4.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.8 | GO:0010842 | retina layer formation(GO:0010842) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 18.3 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 2.2 | 26.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.3 | 24.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.8 | 4.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.8 | 4.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 4.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 3.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 10.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 6.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 2.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 7.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 4.7 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 2.0 | 26.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.7 | 8.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.6 | 4.9 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.0 | 6.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.8 | 4.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 4.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.5 | 2.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.4 | 3.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.4 | 11.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 8.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.3 | 2.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.3 | 24.8 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.2 | 3.6 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 6.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.7 | GO:0030283 | testosterone dehydrogenase [NAD(P)] activity(GO:0030283) |
| 0.1 | 1.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 0.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 4.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 5.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 2.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 9.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 11.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 43.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.7 | 26.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 4.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.5 | 8.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.3 | 4.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 8.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 2.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 6.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 3.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 3.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 3.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 4.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |