Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
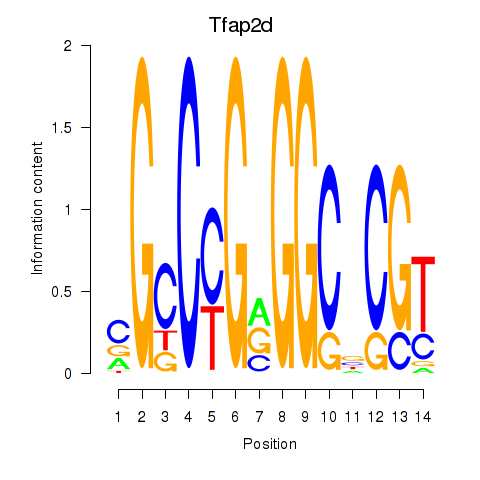
Results for Tfap2d
Z-value: 0.58
Transcription factors associated with Tfap2d
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2d
|
ENSRNOG00000011642 | transcription factor AP-2 delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2d | rn6_v1_chr9_+_25304876_25304876 | 0.00 | 1.0e+00 | Click! |
Activity profile of Tfap2d motif
Sorted Z-values of Tfap2d motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_54734385 | 21.93 |
ENSRNOT00000005023
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr10_+_86340940 | 20.65 |
ENSRNOT00000073486
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr3_-_160301552 | 20.45 |
ENSRNOT00000014498
|
Rims4
|
regulating synaptic membrane exocytosis 4 |
| chr9_+_99998275 | 17.57 |
ENSRNOT00000074395
|
Gpc1
|
glypican 1 |
| chr1_-_270472866 | 13.19 |
ENSRNOT00000015979
|
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr2_+_123791335 | 10.84 |
ENSRNOT00000077096
|
Adad1
|
adenosine deaminase domain containing 1 |
| chr2_+_123790915 | 10.77 |
ENSRNOT00000023282
|
Adad1
|
adenosine deaminase domain containing 1 |
| chr5_+_147714163 | 10.20 |
ENSRNOT00000012663
|
Marcksl1
|
MARCKS-like 1 |
| chr3_+_111545007 | 10.16 |
ENSRNOT00000007247
|
Itpka
|
inositol-trisphosphate 3-kinase A |
| chrX_+_127562660 | 9.78 |
ENSRNOT00000029031
ENSRNOT00000010367 |
Gria3
|
glutamate ionotropic receptor AMPA type subunit 3 |
| chr19_-_11669578 | 9.11 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr4_+_70903253 | 8.32 |
ENSRNOT00000019720
|
Ephb6
|
Eph receptor B6 |
| chr16_-_59366824 | 8.21 |
ENSRNOT00000015062
|
RGD1304810
|
similar to 6430573F11Rik protein |
| chr10_-_98018014 | 8.12 |
ENSRNOT00000005367
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr16_-_21274688 | 7.63 |
ENSRNOT00000027953
|
Tssk6
|
testis-specific serine kinase 6 |
| chr15_+_344685 | 7.31 |
ENSRNOT00000065542
ENSRNOT00000066928 |
Kcnma1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr16_+_26906716 | 6.43 |
ENSRNOT00000064297
|
Cpe
|
carboxypeptidase E |
| chr1_+_263186235 | 5.69 |
ENSRNOT00000021876
|
Cnnm1
|
cyclin and CBS domain divalent metal cation transport mediator 1 |
| chr7_+_116745061 | 5.41 |
ENSRNOT00000076174
|
Rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr9_-_66845403 | 5.40 |
ENSRNOT00000041546
|
Ica1l
|
islet cell autoantigen 1-like |
| chr20_+_13680716 | 5.32 |
ENSRNOT00000029458
|
LOC103694876
|
SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 |
| chr6_+_107485249 | 5.16 |
ENSRNOT00000080859
|
Acot1
|
acyl-CoA thioesterase 1 |
| chr2_+_165486910 | 5.12 |
ENSRNOT00000012982
|
LOC100362176
|
hypothetical protein LOC100362176 |
| chr10_+_13836128 | 5.09 |
ENSRNOT00000012720
|
Pgp
|
phosphoglycolate phosphatase |
| chr3_+_152143811 | 5.02 |
ENSRNOT00000026578
|
LOC100911109
|
sperm-associated antigen 4 protein-like |
| chr8_+_61659445 | 4.94 |
ENSRNOT00000023831
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr8_-_69807826 | 4.90 |
ENSRNOT00000014071
|
Dis3l
|
DIS3-like exosome 3'-5' exoribonuclease |
| chr1_+_143036218 | 4.90 |
ENSRNOT00000025797
|
Pde8a
|
phosphodiesterase 8A |
| chr16_+_4469468 | 4.85 |
ENSRNOT00000021164
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr19_+_25095089 | 4.67 |
ENSRNOT00000041717
|
Prkaca
|
protein kinase cAMP-activated catalytic subunit alpha |
| chr1_-_141349881 | 4.51 |
ENSRNOT00000021109
|
Rhcg
|
Rh family, C glycoprotein |
| chr3_+_176087700 | 4.32 |
ENSRNOT00000073679
|
Mrgbp
|
MRG domain binding protein |
| chr3_-_141305471 | 4.27 |
ENSRNOT00000016915
|
Nkx2-4
|
NK2 homeobox 4 |
| chr16_-_55002322 | 4.24 |
ENSRNOT00000036092
|
Zdhhc2
|
zinc finger, DHHC-type containing 2 |
| chr1_+_101554642 | 4.20 |
ENSRNOT00000028474
|
Bcat2
|
branched chain amino acid transaminase 2 |
| chr15_+_344360 | 4.18 |
ENSRNOT00000077671
|
Kcnma1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr17_+_11953552 | 4.15 |
ENSRNOT00000090782
|
Ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr2_-_186245163 | 4.04 |
ENSRNOT00000089339
|
Dclk2
|
doublecortin-like kinase 2 |
| chr19_-_55300403 | 4.03 |
ENSRNOT00000018591
|
Rnf166
|
ring finger protein 166 |
| chr10_-_16752205 | 3.86 |
ENSRNOT00000079462
|
Crebrf
|
CREB3 regulatory factor |
| chr5_+_127404450 | 3.77 |
ENSRNOT00000017575
|
Lrp8
|
LDL receptor related protein 8 |
| chr1_-_220480132 | 3.73 |
ENSRNOT00000027421
|
Cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr3_-_12029877 | 3.70 |
ENSRNOT00000022327
|
Slc2a8
|
solute carrier family 2 member 8 |
| chr1_+_5448958 | 3.68 |
ENSRNOT00000061930
|
Epm2a
|
epilepsy, progressive myoclonus type 2A |
| chr6_+_93740586 | 3.68 |
ENSRNOT00000011466
|
Dact1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr4_-_113610243 | 3.62 |
ENSRNOT00000008813
|
Hk2
|
hexokinase 2 |
| chr8_+_115179893 | 3.59 |
ENSRNOT00000017469
|
Rrp9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr19_+_34139997 | 3.57 |
ENSRNOT00000017867
|
Arhgap10
|
Rho GTPase activating protein 10 |
| chr14_+_96300596 | 3.41 |
ENSRNOT00000071967
|
LOC302228
|
similar to Spindlin-like protein 2 (SPIN-2) |
| chr16_+_61954590 | 3.36 |
ENSRNOT00000017883
|
Rbpms
|
RNA binding protein with multiple splicing |
| chr5_-_156689258 | 3.25 |
ENSRNOT00000065497
|
Pink1
|
PTEN induced putative kinase 1 |
| chr11_+_87549059 | 3.01 |
ENSRNOT00000002567
|
Ccdc74a
|
coiled-coil domain containing 74A |
| chr1_+_72580424 | 2.97 |
ENSRNOT00000022977
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr4_+_157511642 | 2.93 |
ENSRNOT00000065846
|
Pianp
|
PILR alpha associated neural protein |
| chr3_-_11317049 | 2.89 |
ENSRNOT00000030558
|
Swi5
|
SWI5 homologous recombination repair protein |
| chr10_+_86711240 | 2.86 |
ENSRNOT00000012812
|
Msl1
|
male specific lethal 1 homolog |
| chrX_+_63343724 | 2.71 |
ENSRNOT00000076530
ENSRNOT00000008558 |
Klhl15
|
kelch-like family member 15 |
| chr19_-_25134055 | 2.68 |
ENSRNOT00000007519
|
Misp3
|
MISP family member 3 |
| chr4_+_145580799 | 2.66 |
ENSRNOT00000013727
|
Vhl
|
von Hippel-Lindau tumor suppressor |
| chr3_-_15433252 | 2.58 |
ENSRNOT00000008817
|
Lhx6
|
LIM homeobox 6 |
| chrX_+_63343546 | 2.57 |
ENSRNOT00000076315
|
Klhl15
|
kelch-like family member 15 |
| chr18_-_18079560 | 2.51 |
ENSRNOT00000072093
|
AABR07031533.1
|
|
| chr10_-_68142864 | 2.44 |
ENSRNOT00000004609
|
Myo1d
|
myosin ID |
| chr3_+_175924522 | 2.40 |
ENSRNOT00000086626
|
NEWGENE_1308612
|
MRG/MORF4L binding protein |
| chr6_+_137243185 | 2.35 |
ENSRNOT00000030879
|
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr2_+_195684210 | 2.28 |
ENSRNOT00000028316
|
mrpl9
|
mitochondrial ribosomal protein L9 |
| chr9_+_66716969 | 2.24 |
ENSRNOT00000031261
|
Fam117b
|
family with sequence similarity 117, member B |
| chr6_+_107596782 | 2.24 |
ENSRNOT00000076882
|
Dnal1
|
dynein, axonemal, light chain 1 |
| chr20_-_6960481 | 2.20 |
ENSRNOT00000093172
|
Mtch1
|
mitochondrial carrier 1 |
| chr14_-_11198194 | 2.19 |
ENSRNOT00000003083
|
Enoph1
|
enolase-phosphatase 1 |
| chr2_-_186245342 | 2.19 |
ENSRNOT00000057062
ENSRNOT00000022292 |
Dclk2
|
doublecortin-like kinase 2 |
| chr16_-_81756654 | 2.11 |
ENSRNOT00000026653
|
Cul4a
|
cullin 4A |
| chr19_+_24747178 | 2.03 |
ENSRNOT00000038879
ENSRNOT00000079106 |
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr19_-_25801526 | 2.00 |
ENSRNOT00000003884
|
Nacc1
|
nucleus accumbens associated 1 |
| chr1_-_164590562 | 1.97 |
ENSRNOT00000024157
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr10_+_91830654 | 1.95 |
ENSRNOT00000005176
|
Wnt3
|
wingless-type MMTV integration site family, member 3 |
| chr8_+_53678777 | 1.93 |
ENSRNOT00000045944
|
Drd2
|
dopamine receptor D2 |
| chr1_-_80515694 | 1.90 |
ENSRNOT00000075144
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chr3_+_79678201 | 1.81 |
ENSRNOT00000087604
ENSRNOT00000079709 |
Mtch2
|
mitochondrial carrier 2 |
| chr13_+_51795867 | 1.80 |
ENSRNOT00000006747
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chr17_+_36334147 | 1.78 |
ENSRNOT00000050261
|
E2f3
|
E2F transcription factor 3 |
| chr10_-_14061703 | 1.76 |
ENSRNOT00000017917
|
Gfer
|
growth factor, augmenter of liver regeneration |
| chr8_-_60830448 | 1.75 |
ENSRNOT00000022419
|
Tspan3
|
tetraspanin 3 |
| chr11_-_34598102 | 1.73 |
ENSRNOT00000068743
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr12_+_50090464 | 1.71 |
ENSRNOT00000032193
|
Sez6l
|
seizure related 6 homolog like |
| chr6_+_137824213 | 1.70 |
ENSRNOT00000056880
|
Pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr5_-_156689415 | 1.69 |
ENSRNOT00000083474
|
Pink1
|
PTEN induced putative kinase 1 |
| chr1_-_226501920 | 1.65 |
ENSRNOT00000050716
|
Lrrc10b
|
leucine rich repeat containing 10B |
| chr11_-_34598275 | 1.64 |
ENSRNOT00000077233
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr10_-_85222861 | 1.61 |
ENSRNOT00000031045
|
Npepps
|
aminopeptidase puromycin sensitive |
| chr9_+_53906073 | 1.60 |
ENSRNOT00000017813
|
Nab1
|
Ngfi-A binding protein 1 |
| chr1_+_214317965 | 1.60 |
ENSRNOT00000078754
|
Tmem80
|
transmembrane protein 80 |
| chr14_-_12387102 | 1.57 |
ENSRNOT00000038872
|
Bmp3
|
bone morphogenetic protein 3 |
| chr3_+_152933771 | 1.45 |
ENSRNOT00000027539
|
RGD1307752
|
similar to RIKEN cDNA 1110008F13 |
| chr16_-_62483137 | 1.45 |
ENSRNOT00000020683
|
Purg
|
purine-rich element binding protein G |
| chr5_+_148193710 | 1.42 |
ENSRNOT00000088568
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr3_-_175644988 | 1.31 |
ENSRNOT00000084452
|
Cables2
|
Cdk5 and Abl enzyme substrate 2 |
| chr12_+_15890170 | 1.29 |
ENSRNOT00000001654
|
Gna12
|
G protein subunit alpha 12 |
| chr1_+_214317682 | 1.27 |
ENSRNOT00000032904
|
Tmem80
|
transmembrane protein 80 |
| chr4_-_145147397 | 1.26 |
ENSRNOT00000010347
|
Lhfpl4
|
lipoma HMGIC fusion partner-like 4 |
| chr10_+_61403130 | 1.26 |
ENSRNOT00000092490
ENSRNOT00000077649 |
Ccdc92b
|
coiled-coil domain containing 92B |
| chr3_+_147772165 | 1.26 |
ENSRNOT00000008713
|
Tbc1d20
|
TBC1 domain family, member 20 |
| chr12_-_17486157 | 1.22 |
ENSRNOT00000001745
|
Get4
|
golgi to ER traffic protein 4 |
| chr6_-_98566523 | 1.15 |
ENSRNOT00000077715
|
Ppp2r5e
|
protein phosphatase 2 regulatory subunit B', epsilon |
| chr8_+_116154913 | 1.12 |
ENSRNOT00000081242
|
Cacna2d2
|
calcium voltage-gated channel auxiliary subunit alpha2delta 2 |
| chr11_-_57993548 | 1.03 |
ENSRNOT00000002957
|
Nectin3
|
nectin cell adhesion molecule 3 |
| chr4_-_58250798 | 0.88 |
ENSRNOT00000048436
|
Klf14
|
Kruppel-like factor 14 |
| chr8_+_116154736 | 0.88 |
ENSRNOT00000021218
|
Cacna2d2
|
calcium voltage-gated channel auxiliary subunit alpha2delta 2 |
| chr2_+_52302170 | 0.88 |
ENSRNOT00000081157
|
Paip1
|
poly(A) binding protein interacting protein 1 |
| chr1_+_198690794 | 0.80 |
ENSRNOT00000023999
|
Zfp771
|
zinc finger protein 771 |
| chr8_+_128087345 | 0.69 |
ENSRNOT00000019777
|
Acvr2b
|
activin A receptor type 2B |
| chr13_+_97702097 | 0.68 |
ENSRNOT00000057787
|
LOC103689999
|
saccharopine dehydrogenase-like oxidoreductase |
| chr10_-_59360661 | 0.61 |
ENSRNOT00000036942
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr7_-_139394166 | 0.58 |
ENSRNOT00000082429
|
Vdr
|
vitamin D (1,25- dihydroxyvitamin D3) receptor |
| chr1_+_84470829 | 0.54 |
ENSRNOT00000025472
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr11_-_44292884 | 0.53 |
ENSRNOT00000084060
|
Dcbld2
|
discoidin, CUB and LCCL domain containing 2 |
| chr14_+_10854682 | 0.49 |
ENSRNOT00000091367
ENSRNOT00000003072 ENSRNOT00000033489 |
Sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr1_-_214408549 | 0.47 |
ENSRNOT00000074016
|
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr1_+_1784078 | 0.40 |
ENSRNOT00000020098
|
Lats1
|
large tumor suppressor kinase 1 |
| chr12_+_31934343 | 0.35 |
ENSRNOT00000011181
|
Tmem132d
|
transmembrane protein 132D |
| chr7_-_70498992 | 0.26 |
ENSRNOT00000067774
ENSRNOT00000079327 |
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase type 2 gamma |
| chr19_+_21372163 | 0.25 |
ENSRNOT00000020329
|
Siah1
|
siah E3 ubiquitin protein ligase 1 |
| chr6_+_28571351 | 0.21 |
ENSRNOT00000005389
|
Adcy3
|
adenylate cyclase 3 |
| chr1_-_125967756 | 0.18 |
ENSRNOT00000031488
|
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr1_-_112947399 | 0.11 |
ENSRNOT00000093306
ENSRNOT00000093259 |
Gabra5
|
gamma-aminobutyric acid type A receptor alpha 5 subunit |
| chr3_-_171342646 | 0.06 |
ENSRNOT00000071853
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr9_+_15068399 | 0.00 |
ENSRNOT00000064229
ENSRNOT00000087107 |
Foxp4
|
forkhead box P4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2d
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.6 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 3.8 | 11.5 | GO:0060082 | eye blink reflex(GO:0060082) |
| 3.4 | 20.6 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 2.1 | 8.3 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 1.7 | 6.8 | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) |
| 1.6 | 8.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 1.5 | 4.5 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 1.4 | 8.6 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 1.4 | 4.2 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 1.4 | 21.9 | GO:0030238 | male sex determination(GO:0030238) |
| 1.3 | 3.9 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 1.3 | 6.4 | GO:2000173 | insulin processing(GO:0030070) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 1.3 | 5.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 1.2 | 3.7 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 1.2 | 4.8 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.1 | 5.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 1.0 | 4.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.9 | 3.7 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.9 | 2.7 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.8 | 7.6 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.8 | 8.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.8 | 4.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.5 | 3.8 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.5 | 10.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.5 | 3.7 | GO:0046959 | habituation(GO:0046959) |
| 0.5 | 4.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.5 | 20.4 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.4 | 5.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.4 | 1.3 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.4 | 9.8 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.4 | 6.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.4 | 2.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.3 | 4.9 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.3 | 2.9 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.3 | 1.2 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.3 | 2.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.3 | 1.8 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) hematopoietic stem cell homeostasis(GO:0061484) |
| 0.3 | 1.8 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.2 | 1.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 10.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 2.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 2.0 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.2 | 0.2 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.2 | 0.6 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.2 | 3.9 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 4.9 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.2 | 27.2 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 0.5 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.4 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 3.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 2.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.7 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 0.7 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 2.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 5.1 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.9 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 1.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 2.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 2.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 2.2 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 3.6 | GO:0006364 | rRNA processing(GO:0006364) rRNA metabolic process(GO:0016072) |
| 0.0 | 1.4 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 2.9 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 21.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 1.1 | 11.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.0 | 2.9 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.7 | 2.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.7 | 9.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 4.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.5 | 4.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.5 | 2.0 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.5 | 9.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 3.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 4.9 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.4 | 6.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 3.9 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.4 | 20.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.4 | 3.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.3 | 1.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.3 | 6.7 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.3 | 2.7 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 5.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.2 | 2.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 7.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 17.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.6 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 2.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 3.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 3.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 2.9 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 5.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 3.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 5.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 3.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 6.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 5.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 8.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 3.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 21.9 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 3.5 | 17.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 2.9 | 11.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 2.1 | 8.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 2.0 | 9.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.8 | 21.6 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 1.6 | 4.9 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 1.5 | 9.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 1.4 | 4.2 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 1.2 | 3.7 | GO:2001070 | starch binding(GO:2001070) |
| 1.2 | 4.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 1.1 | 10.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.8 | 5.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.8 | 3.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.8 | 3.8 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.6 | 3.6 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.6 | 1.8 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.6 | 6.4 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.6 | 3.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.5 | 4.9 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.4 | 5.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.4 | 1.3 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.4 | 3.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.3 | 3.7 | GO:0005536 | glucose binding(GO:0005536) |
| 0.3 | 4.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 20.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.2 | 1.9 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 4.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 4.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.2 | 2.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 4.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.6 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 4.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 21.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 3.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 4.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 2.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 2.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 3.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 2.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 7.1 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 4.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 7.7 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 5.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 5.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 10.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 1.3 | 21.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.5 | 7.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.4 | 11.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 12.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 4.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 2.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 20.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.8 | 17.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.5 | 9.8 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.3 | 4.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 6.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 4.9 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 4.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 16.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 3.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 6.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 3.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 4.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 3.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.6 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 1.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |