Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Tfap2b
Z-value: 0.75
Transcription factors associated with Tfap2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2b
|
ENSRNOG00000011823 | transcription factor AP-2 beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2b | rn6_v1_chr9_+_25410669_25410717 | -0.10 | 6.7e-02 | Click! |
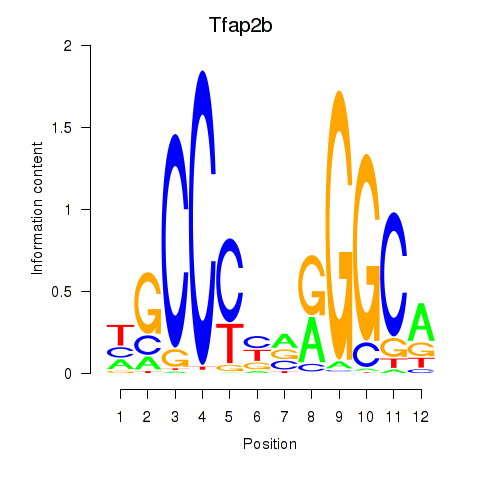
Activity profile of Tfap2b motif
Sorted Z-values of Tfap2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_197858016 | 32.99 |
ENSRNOT00000074778
|
Atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr2_+_189629297 | 24.24 |
ENSRNOT00000049810
|
Dennd4b
|
DENN domain containing 4B |
| chr7_-_116781766 | 23.01 |
ENSRNOT00000010084
|
Mafa
|
MAF bZIP transcription factor A |
| chr17_+_70684539 | 15.39 |
ENSRNOT00000025700
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr7_+_144577465 | 15.17 |
ENSRNOT00000021647
|
Hoxc10
|
homeo box C10 |
| chr17_+_70684134 | 15.05 |
ENSRNOT00000025731
ENSRNOT00000068354 |
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr17_+_70684340 | 14.87 |
ENSRNOT00000051067
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr2_-_238068694 | 12.69 |
ENSRNOT00000079479
ENSRNOT00000078608 ENSRNOT00000090834 |
Npnt
|
nephronectin |
| chr1_+_82480195 | 12.64 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr1_+_87045796 | 12.43 |
ENSRNOT00000027620
|
Lgals7
|
galectin 7 |
| chr4_-_157252104 | 12.35 |
ENSRNOT00000082739
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr10_+_4957326 | 12.22 |
ENSRNOT00000003458
|
Socs1
|
suppressor of cytokine signaling 1 |
| chr10_+_86340940 | 11.37 |
ENSRNOT00000073486
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr6_-_99783047 | 10.93 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr5_-_157199441 | 10.40 |
ENSRNOT00000022559
|
Pla2g2f
|
phospholipase A2, group IIF |
| chr1_+_273563441 | 10.36 |
ENSRNOT00000048691
|
RGD1561333
|
similar to 60S ribosomal protein L8 |
| chr1_-_142183884 | 10.06 |
ENSRNOT00000016032
|
Fes
|
FES proto-oncogene, tyrosine kinase |
| chr20_+_4355175 | 10.06 |
ENSRNOT00000000510
|
Gpsm3
|
G-protein signaling modulator 3 |
| chr19_+_37568113 | 9.79 |
ENSRNOT00000023710
|
Fam65a
|
family with sequence similarity 65, member A |
| chr10_+_84167331 | 9.67 |
ENSRNOT00000010965
|
Hoxb4
|
homeo box B4 |
| chr6_+_10533151 | 9.66 |
ENSRNOT00000020822
|
Rhoq
|
ras homolog family member Q |
| chr14_+_86101277 | 9.64 |
ENSRNOT00000018846
|
Aebp1
|
AE binding protein 1 |
| chr6_+_93281408 | 9.35 |
ENSRNOT00000009610
|
Frmd6
|
FERM domain containing 6 |
| chr1_-_264975132 | 9.31 |
ENSRNOT00000021748
|
Lbx1
|
ladybird homeobox 1 |
| chr3_-_57913999 | 9.22 |
ENSRNOT00000090426
|
Slc25a12
|
solute carrier family 25 member 12 |
| chr1_-_59347472 | 9.20 |
ENSRNOT00000017718
|
Lnpep
|
leucyl and cystinyl aminopeptidase |
| chr7_-_77162148 | 9.20 |
ENSRNOT00000008350
|
Klf10
|
Kruppel-like factor 10 |
| chr19_-_55490426 | 9.06 |
ENSRNOT00000081800
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr7_-_140546908 | 8.96 |
ENSRNOT00000077502
|
Kmt2d
|
lysine methyltransferase 2D |
| chr8_-_72841496 | 8.70 |
ENSRNOT00000057641
ENSRNOT00000040808 ENSRNOT00000085894 ENSRNOT00000024575 ENSRNOT00000048044 ENSRNOT00000024493 |
Tpm1
|
tropomyosin 1, alpha |
| chr19_-_59502161 | 8.43 |
ENSRNOT00000075292
|
Irf2bp2
|
interferon regulatory factor 2 binding protein 2 |
| chr1_+_68436917 | 8.33 |
ENSRNOT00000088586
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr1_+_68436593 | 8.00 |
ENSRNOT00000080325
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr3_-_162581610 | 7.83 |
ENSRNOT00000079324
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr7_-_14303055 | 7.82 |
ENSRNOT00000008963
|
Brd4
|
bromodomain containing 4 |
| chr11_+_85992696 | 7.78 |
ENSRNOT00000084433
|
AABR07034736.1
|
|
| chr20_+_3990820 | 7.69 |
ENSRNOT00000000528
|
Psmb8
|
proteasome subunit beta 8 |
| chr2_+_189169877 | 7.58 |
ENSRNOT00000028225
|
She
|
Src homology 2 domain containing E |
| chr3_+_3389612 | 7.30 |
ENSRNOT00000041984
|
Rpl8
|
ribosomal protein L8 |
| chr3_-_80933283 | 7.29 |
ENSRNOT00000007771
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr9_+_17086475 | 7.24 |
ENSRNOT00000025660
|
Tjap1
|
tight junction associated protein 1 |
| chr3_-_172537877 | 7.03 |
ENSRNOT00000072069
|
Ctsz
|
cathepsin Z |
| chr5_-_152198813 | 7.03 |
ENSRNOT00000082953
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr11_+_82466071 | 6.99 |
ENSRNOT00000036958
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr9_-_20621051 | 6.98 |
ENSRNOT00000015918
|
Tnfrsf21
|
TNF receptor superfamily member 21 |
| chrX_-_38196060 | 6.86 |
ENSRNOT00000006741
ENSRNOT00000006438 |
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr1_+_213991620 | 6.83 |
ENSRNOT00000038691
|
Rplp2
|
ribosomal protein lateral stalk subunit P2 |
| chr1_-_48891130 | 6.75 |
ENSRNOT00000083884
|
AC135026.1
|
|
| chr10_-_56864005 | 6.74 |
ENSRNOT00000033098
ENSRNOT00000091131 |
Alox12
|
arachidonate 12-lipoxygenase, 12S type |
| chr4_-_115354795 | 6.73 |
ENSRNOT00000017691
|
Cd207
|
CD207 molecule |
| chr10_+_39109522 | 6.61 |
ENSRNOT00000010968
|
Irf1
|
interferon regulatory factor 1 |
| chr20_+_3827367 | 6.51 |
ENSRNOT00000079967
|
Rxrb
|
retinoid X receptor beta |
| chr12_+_49761120 | 6.49 |
ENSRNOT00000070961
|
Myo18b
|
myosin XVIIIb |
| chr7_-_63578490 | 6.47 |
ENSRNOT00000007295
|
Rassf3
|
Ras association domain family member 3 |
| chrX_-_114232939 | 6.45 |
ENSRNOT00000042639
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr10_+_17421075 | 6.43 |
ENSRNOT00000047011
|
Stk10
|
serine/threonine kinase 10 |
| chr5_+_151436464 | 6.26 |
ENSRNOT00000012463
|
Map3k6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr13_+_51795867 | 6.23 |
ENSRNOT00000006747
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chr20_+_6973398 | 6.20 |
ENSRNOT00000041665
|
Fgd2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr9_+_93545396 | 5.96 |
ENSRNOT00000025093
|
LOC100359583
|
hypothetical protein LOC100359583 |
| chr1_+_87045335 | 5.92 |
ENSRNOT00000084393
|
Lgals7
|
galectin 7 |
| chr19_+_52647070 | 5.77 |
ENSRNOT00000068389
ENSRNOT00000087857 |
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr10_+_30833914 | 5.76 |
ENSRNOT00000035346
ENSRNOT00000079437 |
Clint1
|
clathrin interactor 1 |
| chr10_+_13145099 | 5.68 |
ENSRNOT00000091287
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr14_+_7618022 | 5.68 |
ENSRNOT00000088508
ENSRNOT00000002819 |
Slc10a6
|
solute carrier family 10 member 6 |
| chr7_-_11777503 | 5.60 |
ENSRNOT00000026220
|
Amh
|
anti-Mullerian hormone |
| chr1_+_101687855 | 5.59 |
ENSRNOT00000028546
ENSRNOT00000091597 |
Dbp
|
D-box binding PAR bZIP transcription factor |
| chr9_+_54212767 | 5.56 |
ENSRNOT00000078073
ENSRNOT00000090026 |
Gls
|
glutaminase |
| chr10_-_62287189 | 5.54 |
ENSRNOT00000004365
|
Wdr81
|
WD repeat domain 81 |
| chr17_-_90627101 | 5.50 |
ENSRNOT00000003349
|
Nid1
|
nidogen 1 |
| chr3_+_172385672 | 5.35 |
ENSRNOT00000090989
|
Gnas
|
GNAS complex locus |
| chr1_+_153589471 | 5.31 |
ENSRNOT00000023205
|
Fzd4
|
frizzled class receptor 4 |
| chr1_-_82580007 | 5.19 |
ENSRNOT00000078668
|
Axl
|
Axl receptor tyrosine kinase |
| chr3_+_155160481 | 5.18 |
ENSRNOT00000021133
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr7_+_27221152 | 5.16 |
ENSRNOT00000031297
|
LOC691921
|
hypothetical protein LOC691921 |
| chr20_-_31072469 | 5.15 |
ENSRNOT00000082448
|
Eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr1_-_222242644 | 5.13 |
ENSRNOT00000050891
|
Vegfb
|
vascular endothelial growth factor B |
| chr1_-_219422268 | 5.06 |
ENSRNOT00000025092
|
Carns1
|
carnosine synthase 1 |
| chr19_-_55423052 | 5.05 |
ENSRNOT00000019528
ENSRNOT00000079641 |
Galns
|
galactosamine (N-acetyl)-6-sulfatase |
| chr5_-_155381732 | 5.04 |
ENSRNOT00000033670
|
Zbtb40
|
zinc finger and BTB domain containing 40 |
| chr13_+_48513008 | 4.97 |
ENSRNOT00000009742
|
Slc26a9
|
solute carrier family 26 member 9 |
| chr7_-_143552588 | 4.97 |
ENSRNOT00000086317
ENSRNOT00000092619 ENSRNOT00000092138 |
Krt78
|
keratin 78 |
| chr19_-_57614558 | 4.91 |
ENSRNOT00000025948
|
RGD1562218
|
similar to RIKEN cDNA 0610039J04 |
| chr5_+_63781801 | 4.89 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr20_-_29511382 | 4.68 |
ENSRNOT00000085026
|
Ddit4
|
DNA-damage-inducible transcript 4 |
| chr15_+_19690194 | 4.63 |
ENSRNOT00000010612
|
Styx
|
serine/threonine/tyrosine interacting protein |
| chrX_+_120859968 | 4.59 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr8_+_118821729 | 4.44 |
ENSRNOT00000028409
|
Setd2
|
SET domain containing 2 |
| chr5_+_59348639 | 4.35 |
ENSRNOT00000084031
ENSRNOT00000060264 |
Reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr6_-_110904288 | 4.34 |
ENSRNOT00000014645
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr4_+_7662019 | 4.33 |
ENSRNOT00000014023
|
Fam126a
|
family with sequence similarity 126, member A |
| chr10_-_85684138 | 4.32 |
ENSRNOT00000017989
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase type 2 beta |
| chr8_+_48805684 | 4.26 |
ENSRNOT00000064041
|
Bcl9l
|
B-cell CLL/lymphoma 9-like |
| chr5_-_159293673 | 3.98 |
ENSRNOT00000009081
|
Padi4
|
peptidyl arginine deiminase 4 |
| chr1_+_177764445 | 3.94 |
ENSRNOT00000037185
|
Rassf10
|
Ras association domain family member 10 |
| chr18_-_40518988 | 3.93 |
ENSRNOT00000004784
|
Fem1c
|
fem-1 homolog C |
| chr1_-_127599257 | 3.85 |
ENSRNOT00000018436
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chr3_-_52998650 | 3.54 |
ENSRNOT00000009988
|
RGD1560831
|
similar to 40S ribosomal protein S3 |
| chr20_+_6049286 | 3.50 |
ENSRNOT00000042143
ENSRNOT00000092566 |
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr16_+_55834458 | 3.47 |
ENSRNOT00000086141
|
AABR07025928.1
|
|
| chr13_-_49169918 | 3.45 |
ENSRNOT00000000036
|
Tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr4_+_157523770 | 3.44 |
ENSRNOT00000055985
ENSRNOT00000023240 |
Zfp384
|
zinc finger protein 384 |
| chr10_-_73787083 | 3.42 |
ENSRNOT00000035280
|
Med13
|
mediator complex subunit 13 |
| chr10_-_35800120 | 3.39 |
ENSRNOT00000004361
|
Maml1
|
mastermind-like transcriptional coactivator 1 |
| chr6_+_26566494 | 3.39 |
ENSRNOT00000079292
|
Gtf3c2
|
general transcription factor IIIC subunit 2 |
| chr13_-_60496511 | 3.39 |
ENSRNOT00000004495
|
Cdc73
|
cell division cycle 73 |
| chr19_+_37252843 | 3.33 |
ENSRNOT00000021145
|
E2f4
|
E2F transcription factor 4 |
| chr4_-_152835182 | 3.33 |
ENSRNOT00000036721
|
B4galnt3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr9_+_46270122 | 2.97 |
ENSRNOT00000031352
|
Cnot11
|
CCR4-NOT transcription complex, subunit 11 |
| chr8_+_128087345 | 2.96 |
ENSRNOT00000019777
|
Acvr2b
|
activin A receptor type 2B |
| chr1_+_47942800 | 2.95 |
ENSRNOT00000079312
|
Wtap
|
Wilms tumor 1 associated protein |
| chrX_+_14498283 | 2.94 |
ENSRNOT00000092143
|
Xk
|
X-linked Kx blood group |
| chr3_-_123702732 | 2.93 |
ENSRNOT00000028859
|
RGD1311739
|
similar to RIKEN cDNA 1700037H04 |
| chr5_+_140692779 | 2.90 |
ENSRNOT00000019101
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr15_+_44411865 | 2.81 |
ENSRNOT00000017566
|
Kctd9
|
potassium channel tetramerization domain containing 9 |
| chr1_+_154377447 | 2.78 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr19_+_57614628 | 2.75 |
ENSRNOT00000026617
|
Gnpat
|
glyceronephosphate O-acyltransferase |
| chr20_+_3823596 | 2.68 |
ENSRNOT00000087670
|
Rxrb
|
retinoid X receptor beta |
| chr12_-_37682964 | 2.59 |
ENSRNOT00000001421
|
RGD1563482
|
similar to hypothetical protein FLJ38663 |
| chr14_-_85398532 | 2.54 |
ENSRNOT00000057361
|
Emid1
|
EMI domain containing 1 |
| chr1_+_78933372 | 2.45 |
ENSRNOT00000029810
|
Ccdc8
|
coiled-coil domain containing 8 |
| chr2_-_243224883 | 2.38 |
ENSRNOT00000014139
|
Dapp1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides 1 |
| chr2_+_116028781 | 2.38 |
ENSRNOT00000089477
|
Phc3
|
polyhomeotic homolog 3 |
| chr16_-_61091169 | 2.37 |
ENSRNOT00000016328
|
Dusp4
|
dual specificity phosphatase 4 |
| chr3_+_46286712 | 2.29 |
ENSRNOT00000085563
|
March7
|
membrane associated ring-CH-type finger 7 |
| chr1_+_47942500 | 2.22 |
ENSRNOT00000025936
|
Wtap
|
Wilms tumor 1 associated protein |
| chr12_-_30501860 | 2.17 |
ENSRNOT00000001227
|
Cct6a
|
chaperonin containing TCP1 subunit 6A |
| chr17_-_14828329 | 2.15 |
ENSRNOT00000022830
|
Barx1
|
BARX homeobox 1 |
| chr5_+_150833819 | 2.12 |
ENSRNOT00000056170
|
Eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr4_+_157647082 | 2.07 |
ENSRNOT00000024938
|
Nop2
|
NOP2 nucleolar protein |
| chr17_-_35037076 | 2.01 |
ENSRNOT00000090946
|
Dusp22
|
dual specificity phosphatase 22 |
| chr8_-_56696968 | 2.00 |
ENSRNOT00000016637
|
Zc3h12c
|
zinc finger CCCH type containing 12C |
| chr3_-_51297852 | 1.99 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chrX_+_68782872 | 1.94 |
ENSRNOT00000075995
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr7_+_141237768 | 1.91 |
ENSRNOT00000091717
|
Aqp2
|
aquaporin 2 |
| chr10_+_110469290 | 1.90 |
ENSRNOT00000054919
|
Foxk2
|
forkhead box K2 |
| chr19_+_46761570 | 1.81 |
ENSRNOT00000058779
|
Wwox
|
WW domain-containing oxidoreductase |
| chr2_+_119197239 | 1.79 |
ENSRNOT00000048030
|
Usp13
|
ubiquitin specific peptidase 13 |
| chr10_+_84119884 | 1.68 |
ENSRNOT00000009951
|
Hoxb9
|
homeo box B9 |
| chr4_-_150616895 | 1.44 |
ENSRNOT00000073562
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr8_-_37126809 | 1.41 |
ENSRNOT00000040783
|
LOC103693040
|
prostate and testis expressed protein 2-like |
| chr15_-_34322115 | 1.38 |
ENSRNOT00000026734
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr7_+_130498199 | 1.29 |
ENSRNOT00000092684
ENSRNOT00000092431 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr1_+_154377247 | 1.27 |
ENSRNOT00000092945
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr3_-_3594475 | 1.20 |
ENSRNOT00000064861
|
RGD1564379
|
RGD1564379 |
| chr1_+_85324079 | 1.19 |
ENSRNOT00000093693
|
Samd4b
|
sterile alpha motif domain containing 4B |
| chr17_+_10676943 | 1.09 |
ENSRNOT00000000116
|
Thoc3
|
THO complex 3 |
| chr12_+_19335841 | 1.07 |
ENSRNOT00000001831
|
Mblac1
|
metallo-beta-lactamase domain containing 1 |
| chr14_-_85982794 | 1.05 |
ENSRNOT00000076874
|
Urgcp
|
upregulator of cell proliferation |
| chr6_+_136042059 | 1.03 |
ENSRNOT00000088784
|
Mark3
|
microtubule affinity regulating kinase 3 |
| chr7_-_141424225 | 1.02 |
ENSRNOT00000080109
ENSRNOT00000084292 |
Cers5
|
ceramide synthase 5 |
| chr1_-_142164007 | 1.01 |
ENSRNOT00000078982
|
Man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr1_-_142164263 | 0.98 |
ENSRNOT00000016281
|
Man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr15_-_28611946 | 0.96 |
ENSRNOT00000016288
|
Supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr8_-_131899023 | 0.90 |
ENSRNOT00000034690
|
Zfp445
|
zinc finger protein 445 |
| chr7_-_26984400 | 0.89 |
ENSRNOT00000013613
|
Txnrd1
|
thioredoxin reductase 1 |
| chr2_+_203200427 | 0.86 |
ENSRNOT00000020566
|
Vtcn1
|
V-set domain containing T cell activation inhibitor 1 |
| chr7_-_119185238 | 0.72 |
ENSRNOT00000007598
|
Foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr1_+_221735517 | 0.70 |
ENSRNOT00000028628
ENSRNOT00000044866 |
Sf1
|
splicing factor 1 |
| chr3_+_126335863 | 0.68 |
ENSRNOT00000028904
|
Bmp2
|
bone morphogenetic protein 2 |
| chr8_+_79606789 | 0.64 |
ENSRNOT00000087114
|
Pygo1
|
pygopus family PHD finger 1 |
| chr8_+_62298358 | 0.63 |
ENSRNOT00000025525
|
Cox5a
|
cytochrome c oxidase subunit 5A |
| chr14_-_86297623 | 0.62 |
ENSRNOT00000067162
ENSRNOT00000081607 ENSRNOT00000085265 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II beta |
| chr13_-_72869396 | 0.51 |
ENSRNOT00000093563
|
RGD1304622
|
similar to 6820428L09 protein |
| chr11_-_83926524 | 0.48 |
ENSRNOT00000041777
ENSRNOT00000040029 |
Eif4g1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_+_264504591 | 0.48 |
ENSRNOT00000050076
|
Pax2
|
paired box 2 |
| chr20_+_3556560 | 0.47 |
ENSRNOT00000085635
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr3_-_147966097 | 0.46 |
ENSRNOT00000009913
|
Defb22
|
defensin beta 22 |
| chr2_+_52301798 | 0.45 |
ENSRNOT00000091387
|
Paip1
|
poly(A) binding protein interacting protein 1 |
| chr12_-_30810964 | 0.42 |
ENSRNOT00000001235
ENSRNOT00000093237 ENSRNOT00000077977 |
Sfswap
|
splicing factor SWAP homolog |
| chr1_+_222519615 | 0.33 |
ENSRNOT00000083585
|
Rcor2
|
REST corepressor 2 |
| chr1_-_242373764 | 0.32 |
ENSRNOT00000076544
|
Fam122a
|
family with sequence similarity 122A |
| chr14_+_64686793 | 0.31 |
ENSRNOT00000005894
ENSRNOT00000036646 |
Adgra3
|
adhesion G protein-coupled receptor A3 |
| chr4_+_140092352 | 0.23 |
ENSRNOT00000008892
|
Setmar
|
SET domain and mariner transposase fusion gene |
| chr4_-_147592699 | 0.05 |
ENSRNOT00000013831
|
Raf1
|
Raf-1 proto-oncogene, serine/threonine kinase |
| chr1_-_198900375 | 0.02 |
ENSRNOT00000024969
|
Zfp689
|
zinc finger protein 689 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.0 | 33.0 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) |
| 4.2 | 12.7 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 4.2 | 12.6 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 3.5 | 24.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 3.2 | 45.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 3.1 | 12.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 3.1 | 9.2 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 2.9 | 8.7 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 2.4 | 4.9 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 2.3 | 9.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 2.0 | 7.8 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 1.9 | 11.4 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 1.9 | 5.6 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 1.9 | 5.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 1.8 | 9.0 | GO:0001555 | oocyte growth(GO:0001555) |
| 1.8 | 5.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 1.7 | 7.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 1.7 | 5.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 1.7 | 8.4 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 1.6 | 9.4 | GO:0003383 | apical constriction(GO:0003383) |
| 1.6 | 6.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.5 | 4.4 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 1.4 | 9.7 | GO:0048539 | bone marrow development(GO:0048539) |
| 1.4 | 6.8 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 1.3 | 6.7 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 1.3 | 6.6 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 1.3 | 5.2 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 1.3 | 5.1 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 1.2 | 7.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 1.2 | 7.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 1.1 | 13.7 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 1.1 | 3.4 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 1.1 | 5.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) progesterone secretion(GO:0042701) |
| 1.0 | 16.3 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 1.0 | 12.2 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.9 | 23.0 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.9 | 7.8 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.8 | 4.0 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.8 | 9.3 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.8 | 10.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.7 | 12.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.6 | 1.9 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.6 | 7.0 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.6 | 2.9 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.6 | 5.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.5 | 6.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.5 | 2.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.5 | 2.9 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.5 | 10.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.4 | 1.8 | GO:1904378 | protein K29-linked deubiquitination(GO:0035523) maintenance of unfolded protein(GO:0036506) protein K6-linked deubiquitination(GO:0044313) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.4 | 9.7 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.4 | 4.3 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.4 | 5.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.4 | 1.2 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.4 | 2.8 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.4 | 4.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.4 | 5.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 10.9 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.3 | 0.7 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) cell-cell signaling involved in cell fate commitment(GO:0045168) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.3 | 2.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.3 | 3.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.3 | 1.3 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 0.3 | 9.2 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.3 | 18.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.3 | 5.0 | GO:0019532 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.3 | 0.6 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.3 | 3.4 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.3 | 0.9 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.3 | 2.2 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.3 | 5.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.3 | 7.2 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.3 | 3.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 0.5 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.2 | 2.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 12.7 | GO:0042130 | negative regulation of T cell proliferation(GO:0042130) |
| 0.2 | 2.0 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 2.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 6.4 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 2.6 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 1.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 5.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 2.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 7.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.9 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 1.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 6.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 3.4 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 6.2 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.5 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 4.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 3.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.2 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 7.7 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 1.3 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 5.1 | GO:1901607 | alpha-amino acid biosynthetic process(GO:1901607) |
| 0.1 | 0.7 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 1.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 4.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 2.1 | GO:0048536 | spleen development(GO:0048536) |
| 0.1 | 0.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 3.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 4.8 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 3.0 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 6.7 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 4.3 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 4.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 9.6 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 2.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.6 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 5.6 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 1.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.7 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 2.9 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 12.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.7 | 15.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 1.5 | 12.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.3 | 7.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.2 | 8.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.1 | 10.9 | GO:0014731 | spectrin(GO:0008091) spectrin-associated cytoskeleton(GO:0014731) |
| 1.0 | 33.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.9 | 7.8 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.9 | 5.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.8 | 3.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.7 | 9.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.7 | 9.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.5 | 10.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.5 | 3.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.5 | 3.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.4 | 2.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.4 | 4.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.3 | 3.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.3 | 1.0 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 5.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 2.3 | GO:0033643 | host cell part(GO:0033643) |
| 0.2 | 7.0 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.2 | 6.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 4.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 5.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.2 | 2.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 2.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 3.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 4.9 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 14.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 11.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 14.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 13.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 1.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 5.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 7.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 9.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 3.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 5.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.5 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 5.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 6.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 13.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 5.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 6.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 5.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 6.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 12.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 75.2 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 10.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 4.6 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 5.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 59.5 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 4.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 3.6 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 33.0 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 4.1 | 45.3 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 3.2 | 9.7 | GO:0032427 | GBD domain binding(GO:0032427) |
| 2.6 | 7.8 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 2.5 | 12.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.9 | 5.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 1.8 | 9.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.7 | 6.7 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 1.5 | 7.3 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 1.4 | 4.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 1.4 | 12.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 1.3 | 9.2 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 1.2 | 7.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.0 | 4.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.9 | 5.4 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.8 | 10.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.8 | 2.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.7 | 2.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.7 | 5.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.6 | 12.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.6 | 5.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.5 | 4.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.5 | 22.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.5 | 10.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.5 | 5.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.5 | 5.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.5 | 7.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.4 | 4.0 | GO:0098748 | clathrin heavy chain binding(GO:0032050) clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 1.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.4 | 2.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.4 | 3.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.4 | 16.7 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.4 | 1.9 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.4 | 5.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.4 | 7.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 2.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.3 | 7.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 5.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 10.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.3 | 5.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 6.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.3 | 7.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.3 | 7.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 7.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 6.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 5.1 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.2 | 7.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 2.6 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 8.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 6.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 4.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 1.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 4.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 8.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 4.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 2.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 3.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 3.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 11.4 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 6.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.9 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 37.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 3.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 12.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.0 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 8.3 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 5.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 10.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 9.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 3.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 23.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 6.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 9.0 | GO:0044212 | regulatory region DNA binding(GO:0000975) transcription regulatory region DNA binding(GO:0044212) |
| 0.1 | 4.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 6.4 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 1.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 1.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 3.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 27.5 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 10.1 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 6.8 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 2.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 4.3 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.6 | 18.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.6 | 45.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.5 | 5.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.5 | 19.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.5 | 5.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.4 | 7.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.4 | 18.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 22.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.3 | 10.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.3 | 7.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.3 | 6.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 7.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.2 | 3.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 6.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 5.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 3.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 6.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 5.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 18.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 15.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 3.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 6.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 1.7 | 31.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 1.4 | 24.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 1.2 | 45.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.9 | 23.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.7 | 12.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.7 | 10.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.6 | 10.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.6 | 10.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.5 | 5.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.5 | 9.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 5.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 3.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.3 | 5.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 8.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 6.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.3 | 7.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.3 | 6.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.3 | 14.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.3 | 2.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.3 | 6.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 5.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 12.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 3.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 1.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 11.0 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.1 | 14.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 3.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 3.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 4.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 17.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 7.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 1.8 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 1.0 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.7 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |