Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Tead3_Tead4
Z-value: 3.09
Transcription factors associated with Tead3_Tead4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
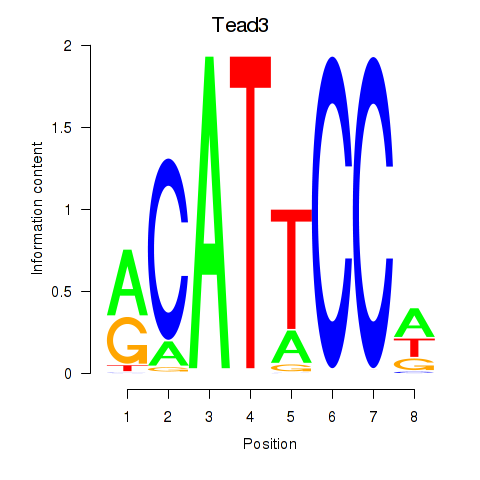
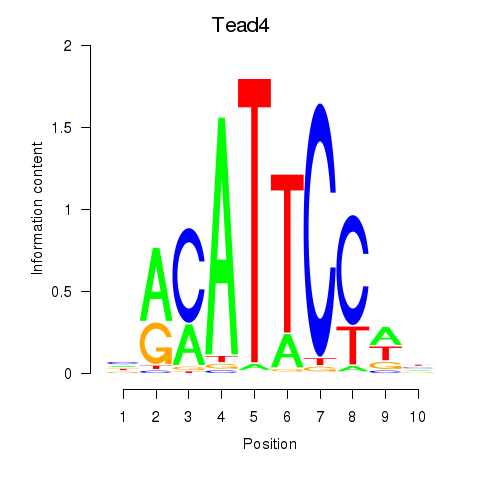
Tead3
|
ENSRNOG00000000506 | TEA domain transcription factor 3 |
|
Tead4
|
ENSRNOG00000005608 | TEA domain transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tead4 | rn6_v1_chr4_-_161587222_161587222 | 0.74 | 2.0e-56 | Click! |
| Tead3 | rn6_v1_chr20_-_7930929_7930929 | 0.06 | 2.5e-01 | Click! |
Activity profile of Tead3_Tead4 motif
Sorted Z-values of Tead3_Tead4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_64952687 | 298.61 |
ENSRNOT00000087892
|
Popdc2
|
popeye domain containing 2 |
| chr1_-_21854763 | 226.27 |
ENSRNOT00000089196
ENSRNOT00000020528 |
Ctgf
|
connective tissue growth factor |
| chr6_-_26624092 | 182.93 |
ENSRNOT00000008113
|
Trim54
|
tripartite motif-containing 54 |
| chr8_-_72841496 | 174.01 |
ENSRNOT00000057641
ENSRNOT00000040808 ENSRNOT00000085894 ENSRNOT00000024575 ENSRNOT00000048044 ENSRNOT00000024493 |
Tpm1
|
tropomyosin 1, alpha |
| chr10_+_92628356 | 156.52 |
ENSRNOT00000072480
|
Myl4
|
myosin, light chain 4 |
| chr5_+_164796185 | 152.86 |
ENSRNOT00000010779
|
Nppb
|
natriuretic peptide B |
| chr8_-_116361343 | 139.40 |
ENSRNOT00000066296
|
Sema3b
|
semaphorin 3B |
| chr13_+_52662996 | 132.16 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr13_-_108841482 | 131.38 |
ENSRNOT00000004632
ENSRNOT00000086775 |
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chrX_+_53053609 | 128.08 |
ENSRNOT00000058357
|
Dmd
|
dystrophin |
| chr10_-_88874528 | 120.43 |
ENSRNOT00000026783
|
Ptrf
|
polymerase I and transcript release factor |
| chr19_+_27404712 | 115.93 |
ENSRNOT00000023657
|
Mylk3
|
myosin light chain kinase 3 |
| chr1_-_252550394 | 111.97 |
ENSRNOT00000083468
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr19_-_56677084 | 110.31 |
ENSRNOT00000024084
|
Acta1
|
actin, alpha 1, skeletal muscle |
| chr8_-_87282156 | 103.98 |
ENSRNOT00000087874
|
Filip1
|
filamin A interacting protein 1 |
| chrX_+_104734082 | 93.31 |
ENSRNOT00000005020
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr9_-_55256340 | 91.23 |
ENSRNOT00000028907
|
Sdpr
|
serum deprivation response |
| chr20_+_26988774 | 88.04 |
ENSRNOT00000090083
|
Mypn
|
myopalladin |
| chr9_+_88357556 | 85.65 |
ENSRNOT00000020669
|
Col4a3
|
collagen type IV alpha 3 chain |
| chr4_+_56711049 | 82.60 |
ENSRNOT00000027237
|
Flnc
|
filamin C |
| chr4_+_56711275 | 80.81 |
ENSRNOT00000088149
|
Flnc
|
filamin C |
| chr8_+_84945444 | 79.09 |
ENSRNOT00000008244
|
Klhl31
|
kelch-like family member 31 |
| chr9_-_80167033 | 78.83 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr9_+_52023295 | 77.66 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr13_+_89524329 | 77.21 |
ENSRNOT00000004279
|
Mpz
|
myelin protein zero |
| chr9_-_80166807 | 75.56 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr8_+_55178289 | 75.20 |
ENSRNOT00000059127
|
Cryab
|
crystallin, alpha B |
| chr10_+_86367596 | 74.40 |
ENSRNOT00000049070
ENSRNOT00000009114 ENSRNOT00000078744 |
Erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr4_-_120559078 | 70.48 |
ENSRNOT00000085730
ENSRNOT00000079575 |
Kbtbd12
|
kelch repeat and BTB domain containing 12 |
| chr8_+_48406260 | 68.50 |
ENSRNOT00000009975
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr9_-_119332967 | 65.36 |
ENSRNOT00000021048
|
Myl12a
|
myosin light chain 12A |
| chr5_+_152533349 | 62.69 |
ENSRNOT00000067524
|
Trim63
|
tripartite motif containing 63 |
| chr4_+_44597123 | 61.80 |
ENSRNOT00000078250
|
Cav1
|
caveolin 1 |
| chr8_-_84632817 | 61.68 |
ENSRNOT00000076942
|
Mlip
|
muscular LMNA-interacting protein |
| chr2_-_257484587 | 60.20 |
ENSRNOT00000083344
|
Nexn
|
nexilin (F actin binding protein) |
| chr13_-_70625842 | 59.35 |
ENSRNOT00000092499
|
Lamc2
|
laminin subunit gamma 2 |
| chrX_+_106774980 | 57.65 |
ENSRNOT00000046091
|
Tceal7
|
transcription elongation factor A like 7 |
| chr1_-_100530183 | 56.68 |
ENSRNOT00000067754
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr4_-_98342887 | 51.69 |
ENSRNOT00000010156
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr4_-_82209933 | 51.24 |
ENSRNOT00000091106
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr4_-_82295470 | 49.92 |
ENSRNOT00000091073
|
Hoxa10
|
homeobox A10 |
| chr6_-_23291568 | 48.97 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr5_+_75392790 | 47.54 |
ENSRNOT00000048457
|
Musk
|
muscle associated receptor tyrosine kinase |
| chr2_-_147693082 | 45.05 |
ENSRNOT00000022507
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr3_+_79860179 | 44.61 |
ENSRNOT00000081160
ENSRNOT00000068444 |
Rapsn
|
receptor-associated protein of the synapse |
| chrX_+_51286737 | 43.89 |
ENSRNOT00000035692
|
Dmd
|
dystrophin |
| chr17_-_14627937 | 43.64 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr11_-_45124423 | 43.52 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr8_-_87158368 | 42.83 |
ENSRNOT00000077071
|
Col12a1
|
collagen type XII alpha 1 chain |
| chr10_-_62699723 | 41.05 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr19_+_61677542 | 40.71 |
ENSRNOT00000014785
|
Itgb1
|
integrin subunit beta 1 |
| chr5_-_152458023 | 40.61 |
ENSRNOT00000031225
|
Cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr9_-_52238564 | 40.36 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr3_+_6430589 | 40.28 |
ENSRNOT00000012334
|
Col5a1
|
collagen type V alpha 1 chain |
| chr10_+_57322331 | 40.26 |
ENSRNOT00000041493
|
Kif1c
|
kinesin family member 1C |
| chr2_-_251532312 | 40.23 |
ENSRNOT00000019501
|
Cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr1_+_198655742 | 39.94 |
ENSRNOT00000023944
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr3_-_177179127 | 39.91 |
ENSRNOT00000021790
|
Sox18
|
SRY box 18 |
| chr19_+_55982740 | 39.74 |
ENSRNOT00000021397
|
Dpep1
|
dipeptidase 1 (renal) |
| chr2_-_261337163 | 39.71 |
ENSRNOT00000030341
|
Tnni3k
|
TNNI3 interacting kinase |
| chr1_+_42121636 | 39.09 |
ENSRNOT00000025616
|
Myct1
|
myc target 1 |
| chr6_+_12362813 | 38.93 |
ENSRNOT00000022370
|
Ston1
|
stonin 1 |
| chr4_+_181286455 | 38.03 |
ENSRNOT00000083633
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr1_+_46835971 | 37.06 |
ENSRNOT00000066798
|
Synj2
|
synaptojanin 2 |
| chr11_+_64522130 | 35.84 |
ENSRNOT00000004315
|
Upk1b
|
uroplakin 1B |
| chr7_+_14037620 | 34.99 |
ENSRNOT00000009606
|
Syde1
|
synapse defective Rho GTPase homolog 1 |
| chr7_-_98197087 | 34.68 |
ENSRNOT00000010484
ENSRNOT00000079961 |
Klhl38
|
kelch-like family member 38 |
| chr15_+_86243148 | 34.54 |
ENSRNOT00000084471
ENSRNOT00000090727 |
Lmo7
|
LIM domain 7 |
| chr2_+_205592182 | 33.95 |
ENSRNOT00000081050
|
Dennd2c
|
DENN domain containing 2C |
| chr15_+_4064706 | 33.93 |
ENSRNOT00000011956
|
Synpo2l
|
synaptopodin 2-like |
| chr8_-_62948720 | 33.78 |
ENSRNOT00000075476
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr3_+_150801289 | 33.63 |
ENSRNOT00000035060
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr4_-_64981384 | 33.14 |
ENSRNOT00000017338
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr13_-_70626252 | 33.02 |
ENSRNOT00000036947
|
Lamc2
|
laminin subunit gamma 2 |
| chr15_-_33218456 | 32.80 |
ENSRNOT00000017753
|
Ajuba
|
ajuba LIM protein |
| chr14_-_6645257 | 32.52 |
ENSRNOT00000002916
|
Pkd2
|
polycystin 2, transient receptor potential cation channel |
| chr19_+_20147037 | 31.66 |
ENSRNOT00000020028
|
Zfp423
|
zinc finger protein 423 |
| chr17_-_84247038 | 31.61 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr9_+_50526811 | 29.54 |
ENSRNOT00000036990
|
RGD1305645
|
similar to RIKEN cDNA 1500015O10 |
| chr18_+_57286322 | 28.75 |
ENSRNOT00000026174
|
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr2_-_219842986 | 27.95 |
ENSRNOT00000055735
|
Agl
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chrX_+_22247088 | 27.65 |
ENSRNOT00000079802
|
Iqsec2
|
IQ motif and Sec7 domain 2 |
| chr20_+_3555135 | 27.49 |
ENSRNOT00000085380
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_-_206222248 | 27.26 |
ENSRNOT00000026075
|
Olfml3
|
olfactomedin-like 3 |
| chr10_-_104358253 | 26.39 |
ENSRNOT00000005942
|
Caskin2
|
cask-interacting protein 2 |
| chr7_-_92996025 | 26.18 |
ENSRNOT00000076327
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr1_+_41590494 | 25.47 |
ENSRNOT00000089984
|
Esr1
|
estrogen receptor 1 |
| chr1_-_261371508 | 25.47 |
ENSRNOT00000019978
|
Avpi1
|
arginine vasopressin-induced 1 |
| chr5_+_113725717 | 25.40 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr19_+_10519493 | 25.37 |
ENSRNOT00000030229
|
Ccdc102a
|
coiled-coil domain containing 102A |
| chr2_-_233743866 | 25.37 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
| chr19_-_55367353 | 25.34 |
ENSRNOT00000091139
|
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr7_-_81592206 | 24.67 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr5_-_157268903 | 24.57 |
ENSRNOT00000022716
|
Pla2g5
|
phospholipase A2, group V |
| chr10_+_15672382 | 24.51 |
ENSRNOT00000027965
|
Rhbdf1
|
rhomboid 5 homolog 1 |
| chr2_-_44907030 | 24.32 |
ENSRNOT00000013979
|
Gpx8
|
glutathione peroxidase 8 |
| chr9_+_16737642 | 24.23 |
ENSRNOT00000061430
|
Srf
|
serum response factor |
| chr4_-_17594598 | 24.17 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr11_+_27208564 | 24.17 |
ENSRNOT00000002158
|
Map3k7cl
|
MAP3K7 C-terminal like |
| chr8_+_111210811 | 23.49 |
ENSRNOT00000011347
|
Amotl2
|
angiomotin like 2 |
| chr1_-_189960073 | 23.39 |
ENSRNOT00000088482
|
Crym
|
crystallin, mu |
| chr16_+_59197367 | 23.33 |
ENSRNOT00000014924
|
Dlc1
|
DLC1 Rho GTPase activating protein |
| chr10_-_107386072 | 23.33 |
ENSRNOT00000004290
|
Timp2
|
TIMP metallopeptidase inhibitor 2 |
| chr1_-_250774105 | 23.12 |
ENSRNOT00000079227
|
Sgms1
|
sphingomyelin synthase 1 |
| chr1_+_80279706 | 22.36 |
ENSRNOT00000047105
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr13_+_52147555 | 22.18 |
ENSRNOT00000084766
|
Lmod1
|
leiomodin 1 |
| chr1_-_18058055 | 21.93 |
ENSRNOT00000020988
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr4_+_29978739 | 21.93 |
ENSRNOT00000011756
|
Ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr1_+_85386470 | 21.91 |
ENSRNOT00000093332
ENSRNOT00000044326 |
Plekhg2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr6_-_99783047 | 21.82 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr6_-_122721496 | 21.33 |
ENSRNOT00000079697
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr6_+_1147012 | 21.17 |
ENSRNOT00000006360
|
Vit
|
vitrin |
| chr7_-_144993652 | 20.25 |
ENSRNOT00000087748
|
Itga5
|
integrin subunit alpha 5 |
| chr8_+_75687100 | 20.21 |
ENSRNOT00000038677
|
Anxa2
|
annexin A2 |
| chr5_+_25349928 | 20.19 |
ENSRNOT00000020826
|
Gem
|
GTP binding protein overexpressed in skeletal muscle |
| chr18_-_55771730 | 20.14 |
ENSRNOT00000026426
|
Smim3
|
small integral membrane protein 3 |
| chr14_-_114692764 | 20.02 |
ENSRNOT00000008210
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr5_+_74649765 | 19.99 |
ENSRNOT00000075952
|
Palm2
|
paralemmin 2 |
| chr10_-_85636902 | 19.96 |
ENSRNOT00000017241
|
Pcgf2
|
polycomb group ring finger 2 |
| chr5_-_158439078 | 19.79 |
ENSRNOT00000025517
|
Klhdc7a
|
kelch domain containing 7A |
| chr10_-_56403188 | 19.65 |
ENSRNOT00000019947
|
Chrnb1
|
cholinergic receptor nicotinic beta 1 subunit |
| chr1_+_218466289 | 19.25 |
ENSRNOT00000017948
|
Mrgprf
|
MAS related GPR family member F |
| chrX_+_104684676 | 19.18 |
ENSRNOT00000083229
|
Tnmd
|
tenomodulin |
| chr16_-_64778486 | 18.75 |
ENSRNOT00000031701
|
Rnf122
|
ring finger protein 122 |
| chr5_+_58393233 | 18.75 |
ENSRNOT00000000142
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr5_+_74941802 | 17.88 |
ENSRNOT00000015576
|
Akap2
|
A-kinase anchoring protein 2 |
| chr20_+_44680449 | 17.88 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chrX_+_156463953 | 17.01 |
ENSRNOT00000079889
|
Flna
|
filamin A |
| chr5_-_139749050 | 16.84 |
ENSRNOT00000056651
|
Nfyc
|
nuclear transcription factor Y subunit gamma |
| chr6_-_21135880 | 16.70 |
ENSRNOT00000051239
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chr1_+_44311513 | 16.68 |
ENSRNOT00000065386
|
Tiam2
|
T-cell lymphoma invasion and metastasis 2 |
| chr2_+_187737770 | 16.09 |
ENSRNOT00000036143
ENSRNOT00000092611 ENSRNOT00000092620 |
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr16_+_83161880 | 16.00 |
ENSRNOT00000080793
|
1700016D06Rik
|
RIKEN cDNA 1700016D06 gene |
| chr3_+_139894331 | 15.74 |
ENSRNOT00000064695
|
Rin2
|
Ras and Rab interactor 2 |
| chr1_+_264768105 | 15.72 |
ENSRNOT00000086766
ENSRNOT00000048843 |
Lzts2
|
leucine zipper tumor suppressor 2 |
| chr5_-_102743417 | 15.22 |
ENSRNOT00000067389
|
Bnc2
|
basonuclin 2 |
| chr1_-_2846200 | 14.91 |
ENSRNOT00000017688
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr1_+_199163086 | 14.70 |
ENSRNOT00000025646
|
Ctf1
|
cardiotrophin 1 |
| chr12_+_22451596 | 14.01 |
ENSRNOT00000072838
|
Trip6
|
thyroid hormone receptor interactor 6 |
| chr10_+_104523996 | 13.72 |
ENSRNOT00000065339
ENSRNOT00000086747 |
Itgb4
|
integrin subunit beta 4 |
| chr17_+_70010794 | 13.70 |
ENSRNOT00000023945
|
Net1
|
neuroepithelial cell transforming 1 |
| chr13_-_107886476 | 13.65 |
ENSRNOT00000077282
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr14_-_60276794 | 13.37 |
ENSRNOT00000048509
|
Slc34a2
|
solute carrier family 34 member 2 |
| chr14_-_8432195 | 13.36 |
ENSRNOT00000089800
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chrX_-_77675487 | 13.00 |
ENSRNOT00000042799
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr1_-_189911571 | 12.83 |
ENSRNOT00000080996
ENSRNOT00000088536 |
Zp2
|
zona pellucida glycoprotein 2 |
| chr1_+_41323194 | 12.81 |
ENSRNOT00000026350
|
Esr1
|
estrogen receptor 1 |
| chr15_-_8914501 | 12.73 |
ENSRNOT00000008752
|
Thrb
|
thyroid hormone receptor beta |
| chr7_+_34952011 | 12.17 |
ENSRNOT00000077666
|
Fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr7_-_144319804 | 12.07 |
ENSRNOT00000070855
|
LOC100912282
|
calcium-binding and coiled-coil domain-containing protein 1-like |
| chr19_+_568287 | 11.79 |
ENSRNOT00000016419
|
Cdh16
|
cadherin 16 |
| chr1_-_131460473 | 11.73 |
ENSRNOT00000084336
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr5_+_171478555 | 11.64 |
ENSRNOT00000091417
ENSRNOT00000000169 |
LOC100911486
|
multiple epidermal growth factor-like domains protein 6-like |
| chr19_-_49448072 | 11.49 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr12_+_40895515 | 10.92 |
ENSRNOT00000046323
|
Ptpn11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr3_+_160945359 | 10.79 |
ENSRNOT00000019761
|
Pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr2_-_25235275 | 10.74 |
ENSRNOT00000061580
|
F2rl1
|
F2R like trypsin receptor 1 |
| chr5_+_159845774 | 10.57 |
ENSRNOT00000012328
|
Epha2
|
Eph receptor A2 |
| chr10_-_90657185 | 10.46 |
ENSRNOT00000003685
|
Ccdc43
|
coiled-coil domain containing 43 |
| chr3_+_75354237 | 10.39 |
ENSRNOT00000050634
|
Olr557
|
olfactory receptor 557 |
| chr3_+_2676296 | 10.35 |
ENSRNOT00000020497
|
Clic3
|
chloride intracellular channel 3 |
| chr10_-_59049482 | 10.28 |
ENSRNOT00000078272
|
Spns2
|
spinster homolog 2 |
| chr6_-_86822094 | 10.23 |
ENSRNOT00000006531
|
Fkbp3
|
FK506 binding protein 3 |
| chr10_+_74874130 | 10.06 |
ENSRNOT00000076050
ENSRNOT00000044226 ENSRNOT00000085052 ENSRNOT00000076196 |
Sept4
|
septin 4 |
| chr3_-_74906989 | 10.05 |
ENSRNOT00000071001
|
Olr545
|
olfactory receptor 545 |
| chr2_+_41157823 | 9.80 |
ENSRNOT00000066384
|
Pde4d
|
phosphodiesterase 4D |
| chr10_+_74413989 | 9.73 |
ENSRNOT00000036098
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chrX_+_77119911 | 9.73 |
ENSRNOT00000080141
|
Atp7a
|
ATPase copper transporting alpha |
| chr2_-_193136520 | 9.12 |
ENSRNOT00000042142
|
Kprp
|
keratinocyte proline-rich protein |
| chr13_+_34498749 | 8.62 |
ENSRNOT00000093127
|
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr3_-_94767239 | 8.42 |
ENSRNOT00000056808
|
Qser1
|
glutamine and serine rich 1 |
| chr7_-_143016040 | 8.22 |
ENSRNOT00000029697
|
Krt80
|
keratin 80 |
| chr9_+_54212767 | 8.18 |
ENSRNOT00000078073
ENSRNOT00000090026 |
Gls
|
glutaminase |
| chr1_-_31055453 | 8.15 |
ENSRNOT00000031083
|
Soga3
|
SOGA family member 3 |
| chr2_+_41442241 | 8.11 |
ENSRNOT00000067546
|
Pde4d
|
phosphodiesterase 4D |
| chr19_-_11341863 | 7.81 |
ENSRNOT00000025694
|
Mt4
|
metallothionein 4 |
| chr7_-_34951644 | 7.78 |
ENSRNOT00000030015
|
Vezt
|
vezatin, adherens junctions transmembrane protein |
| chr1_-_215553451 | 7.52 |
ENSRNOT00000027407
|
Ctsd
|
cathepsin D |
| chr6_-_136436620 | 7.40 |
ENSRNOT00000067118
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chr20_-_3439983 | 7.40 |
ENSRNOT00000080822
ENSRNOT00000001099 |
Ier3
|
immediate early response 3 |
| chr5_-_135285311 | 7.39 |
ENSRNOT00000088261
ENSRNOT00000085235 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr3_+_111825162 | 7.31 |
ENSRNOT00000066685
|
Pla2g4b
|
phospholipase A2 group IVB |
| chr19_+_43163562 | 7.13 |
ENSRNOT00000048900
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr5_+_169274785 | 7.12 |
ENSRNOT00000082586
|
Plekhg5
|
pleckstrin homology and RhoGEF domain containing G5 |
| chr16_+_71738718 | 7.07 |
ENSRNOT00000022097
|
Plekha2
|
pleckstrin homology domain containing A2 |
| chr1_+_88078350 | 6.67 |
ENSRNOT00000048677
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr2_-_219458271 | 6.32 |
ENSRNOT00000064735
|
Cdc14a
|
cell division cycle 14A |
| chr3_+_75322884 | 6.08 |
ENSRNOT00000066249
|
Olr556
|
olfactory receptor 556 |
| chr6_-_136436818 | 6.07 |
ENSRNOT00000082600
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chrX_-_26376467 | 6.07 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr13_+_88265331 | 6.06 |
ENSRNOT00000031190
|
Ccdc190
|
coiled-coil domain containing 190 |
| chr4_+_10020349 | 6.03 |
ENSRNOT00000065039
|
Fbxl13
|
F-box and leucine-rich repeat protein 13 |
| chr11_+_15436269 | 5.98 |
ENSRNOT00000002150
ENSRNOT00000084928 |
Usp25
|
ubiquitin specific peptidase 25 |
| chr19_-_29968424 | 5.96 |
ENSRNOT00000024981
|
Inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr11_-_28900376 | 5.69 |
ENSRNOT00000061606
|
Krtap16-5
|
keratin associated protein 16-5 |
| chr4_+_157125998 | 5.68 |
ENSRNOT00000078349
|
C1r
|
complement C1r |
| chr15_+_105640097 | 5.44 |
ENSRNOT00000014300
|
Mbnl2
|
muscleblind-like splicing regulator 2 |
| chr1_-_168471161 | 5.43 |
ENSRNOT00000055227
|
Olr94
|
olfactory receptor 94 |
| chr2_-_173800761 | 5.40 |
ENSRNOT00000039524
|
Wdr49
|
WD repeat domain 49 |
| chr18_-_28516209 | 5.39 |
ENSRNOT00000072580
|
Ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tead3_Tead4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 99.5 | 298.6 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 58.0 | 174.0 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 57.3 | 172.0 | GO:0016203 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 45.3 | 226.3 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 44.5 | 222.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 44.1 | 132.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 38.6 | 154.4 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 25.9 | 77.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 23.2 | 115.9 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 21.8 | 152.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 20.6 | 61.8 | GO:1903598 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) regulation of inward rectifier potassium channel activity(GO:1901979) positive regulation of gap junction assembly(GO:1903598) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 20.2 | 80.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 16.7 | 50.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 15.8 | 47.5 | GO:2000541 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 14.9 | 74.4 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 14.9 | 44.6 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 13.6 | 40.7 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 13.3 | 39.9 | GO:0060214 | endocardium formation(GO:0060214) |
| 12.5 | 62.7 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 12.4 | 148.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 10.8 | 32.5 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) renal artery morphogenesis(GO:0061441) |
| 10.5 | 31.6 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 10.4 | 93.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 9.6 | 38.3 | GO:1990375 | baculum development(GO:1990375) |
| 9.4 | 75.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 8.3 | 141.9 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 8.2 | 24.6 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 8.1 | 24.2 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 8.0 | 40.2 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 6.7 | 20.2 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 6.6 | 85.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 6.3 | 25.4 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 6.3 | 56.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 6.0 | 17.9 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 5.9 | 11.7 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 5.6 | 45.0 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 5.6 | 78.0 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 5.5 | 49.6 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 5.4 | 10.7 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 4.9 | 14.7 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 4.8 | 101.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 4.8 | 19.2 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 4.7 | 14.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 4.6 | 156.5 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 4.5 | 13.6 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 4.5 | 148.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 4.2 | 29.5 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 4.1 | 44.6 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 4.0 | 68.5 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 3.7 | 14.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 3.7 | 22.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 3.6 | 25.3 | GO:0033634 | positive regulation of integrin activation(GO:0033625) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 3.5 | 66.4 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 3.4 | 17.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905031) |
| 3.3 | 23.3 | GO:1905049 | negative regulation of metallopeptidase activity(GO:1905049) |
| 3.3 | 23.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 3.3 | 183.1 | GO:0048747 | muscle fiber development(GO:0048747) |
| 3.2 | 9.7 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 3.2 | 12.8 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 2.9 | 61.7 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 2.9 | 35.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 2.7 | 10.9 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 2.7 | 8.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 2.7 | 29.4 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 2.6 | 37.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 2.6 | 28.7 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 2.5 | 20.0 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 2.4 | 33.6 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 2.2 | 39.7 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 2.2 | 13.0 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 2.1 | 23.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 2.1 | 12.7 | GO:0008050 | female courtship behavior(GO:0008050) |
| 2.1 | 39.7 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 2.0 | 10.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 2.0 | 59.6 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 2.0 | 13.7 | GO:0051451 | myoblast migration(GO:0051451) |
| 1.9 | 49.9 | GO:0060065 | uterus development(GO:0060065) |
| 1.9 | 19.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.9 | 13.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 1.9 | 24.2 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 1.8 | 5.4 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 1.7 | 13.7 | GO:0035878 | nail development(GO:0035878) trophoblast cell migration(GO:0061450) |
| 1.7 | 5.0 | GO:1904683 | regulation of metalloendopeptidase activity(GO:1904683) positive regulation of metalloendopeptidase activity(GO:1904685) |
| 1.6 | 22.4 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 1.4 | 4.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 1.4 | 15.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 1.4 | 28.0 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 1.3 | 10.3 | GO:0048069 | eye pigmentation(GO:0048069) |
| 1.3 | 5.0 | GO:1903576 | response to L-arginine(GO:1903576) |
| 1.1 | 31.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 1.1 | 40.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 1.1 | 42.8 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 1.0 | 43.6 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 1.0 | 3.9 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 1.0 | 12.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.9 | 17.9 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.9 | 7.4 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.9 | 3.6 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.9 | 2.7 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.9 | 27.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.9 | 2.6 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.8 | 7.4 | GO:0051198 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.8 | 3.2 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 0.8 | 3.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.8 | 58.5 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.7 | 11.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.7 | 1.4 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.7 | 15.2 | GO:0043586 | tongue development(GO:0043586) |
| 0.6 | 10.9 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.6 | 3.0 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.6 | 34.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.5 | 48.6 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.5 | 2.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.5 | 6.0 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.5 | 30.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.5 | 5.3 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.5 | 2.9 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.5 | 3.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 59.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.4 | 7.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.4 | 20.2 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.4 | 7.1 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.3 | 3.1 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.3 | 25.5 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.3 | 52.2 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.3 | 6.7 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.3 | 7.5 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.3 | 4.7 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.3 | 24.5 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.3 | 20.0 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.3 | 2.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 6.0 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.3 | 3.1 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.3 | 24.3 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.3 | 1.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 2.8 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 4.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 10.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 2.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 15.8 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.2 | 2.0 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.1 | 16.7 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.1 | 3.8 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.1 | 4.4 | GO:0048536 | spleen development(GO:0048536) |
| 0.1 | 5.7 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 4.3 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 29.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 12.3 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 3.1 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 4.3 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.1 | 1.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 2.5 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 2.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 9.1 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.1 | 8.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.4 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 3.7 | GO:0030198 | extracellular matrix organization(GO:0030198) |
| 0.0 | 33.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.9 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 33.0 | 132.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 25.7 | 154.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 24.9 | 174.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 23.1 | 92.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 22.4 | 112.0 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 20.2 | 80.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 13.6 | 40.7 | GO:0034667 | integrin alpha2-beta1 complex(GO:0034666) integrin alpha3-beta1 complex(GO:0034667) |
| 12.5 | 75.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 10.7 | 42.8 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 10.7 | 85.6 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 9.7 | 77.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 9.1 | 172.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) filopodium membrane(GO:0031527) glycoprotein complex(GO:0090665) |
| 7.9 | 110.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 7.4 | 37.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 6.9 | 61.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 6.4 | 122.0 | GO:0016460 | myosin II complex(GO:0016460) |
| 6.2 | 74.4 | GO:0043219 | lateral loop(GO:0043219) |
| 5.5 | 21.9 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 5.3 | 239.2 | GO:0031672 | A band(GO:0031672) |
| 5.1 | 163.4 | GO:0043034 | costamere(GO:0043034) |
| 4.6 | 32.5 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 4.4 | 21.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 4.0 | 20.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 3.9 | 35.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 3.8 | 38.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 3.8 | 424.6 | GO:0030018 | Z disc(GO:0030018) |
| 3.8 | 206.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 3.7 | 44.6 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 2.9 | 23.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 2.9 | 20.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 2.8 | 19.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 2.7 | 53.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 2.5 | 93.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 2.4 | 28.8 | GO:0044754 | autolysosome(GO:0044754) |
| 2.2 | 8.6 | GO:0031592 | centrosomal corona(GO:0031592) |
| 2.1 | 266.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 1.8 | 25.4 | GO:0031983 | vesicle lumen(GO:0031983) |
| 1.8 | 25.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 1.8 | 133.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 1.7 | 13.6 | GO:0097449 | astrocyte projection(GO:0097449) |
| 1.7 | 13.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.6 | 65.3 | GO:0016459 | myosin complex(GO:0016459) |
| 1.5 | 114.6 | GO:0005901 | caveola(GO:0005901) |
| 1.3 | 7.8 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 1.2 | 21.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.0 | 16.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.0 | 19.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 1.0 | 40.1 | GO:0030016 | myofibril(GO:0030016) |
| 0.9 | 20.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.9 | 47.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.8 | 8.4 | GO:0097227 | septin complex(GO:0031105) sperm annulus(GO:0097227) |
| 0.8 | 28.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.8 | 3.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.7 | 9.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.7 | 4.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.6 | 57.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.6 | 14.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.6 | 34.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.6 | 8.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.6 | 198.0 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.6 | 4.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.5 | 4.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.5 | 78.6 | GO:0005938 | cell cortex(GO:0005938) |
| 0.5 | 35.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.5 | 50.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.5 | 15.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.5 | 2.9 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.5 | 27.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.5 | 44.2 | GO:0016605 | PML body(GO:0016605) |
| 0.4 | 107.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.3 | 5.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 11.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 0.9 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.3 | 0.9 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.3 | 21.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.3 | 60.1 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.2 | 29.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.2 | 66.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.2 | 15.7 | GO:0030496 | midbody(GO:0030496) |
| 0.2 | 4.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 88.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.2 | 7.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 23.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.2 | 37.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 18.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 12.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 4.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 2.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 137.8 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 5.5 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 33.0 | 132.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 29.0 | 115.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 17.2 | 154.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 14.9 | 74.4 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 14.0 | 280.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 13.6 | 40.7 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 13.2 | 39.7 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 13.2 | 39.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 11.7 | 446.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 11.1 | 177.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 8.4 | 117.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 7.7 | 316.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 7.7 | 38.3 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 7.1 | 205.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 7.0 | 62.7 | GO:0031432 | titin binding(GO:0031432) |
| 6.9 | 75.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 6.6 | 163.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 5.7 | 17.0 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 5.3 | 159.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 5.0 | 44.6 | GO:0043495 | protein anchor(GO:0043495) |
| 4.9 | 14.7 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 4.7 | 28.0 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 4.6 | 37.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 4.6 | 23.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 4.0 | 20.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 3.8 | 53.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 3.6 | 25.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 3.5 | 31.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 3.4 | 75.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 3.4 | 33.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 3.2 | 12.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 3.0 | 36.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 2.9 | 168.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 2.8 | 25.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 2.7 | 32.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 2.7 | 19.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 2.7 | 8.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 2.7 | 10.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 2.7 | 13.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 2.6 | 13.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 2.4 | 24.2 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 2.2 | 20.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 2.1 | 156.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 2.0 | 6.0 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 1.9 | 9.7 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 1.9 | 23.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 1.9 | 23.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 1.9 | 24.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.8 | 5.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 1.8 | 56.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 1.7 | 10.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 1.7 | 26.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 1.7 | 91.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 1.2 | 31.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 1.2 | 122.2 | GO:0019838 | growth factor binding(GO:0019838) |
| 1.2 | 3.6 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 1.2 | 19.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 1.1 | 27.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 1.1 | 25.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 1.1 | 58.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 1.1 | 24.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 1.0 | 8.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.9 | 40.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.9 | 30.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.9 | 95.0 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.9 | 11.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.9 | 21.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.9 | 14.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.9 | 10.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.8 | 2.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.7 | 44.8 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.7 | 4.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.7 | 20.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.7 | 92.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.6 | 21.2 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.6 | 6.1 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.5 | 57.8 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.5 | 2.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.5 | 3.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.5 | 2.9 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.5 | 7.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.4 | 17.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.4 | 112.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.4 | 25.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.4 | 3.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.4 | 0.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.4 | 22.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.4 | 44.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.4 | 6.0 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.3 | 33.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.3 | 32.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.3 | 7.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.3 | 6.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.3 | 2.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 5.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 5.0 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 68.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.2 | 4.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 33.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.2 | 22.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 42.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 16.1 | GO:0005496 | steroid binding(GO:0005496) |
| 0.2 | 4.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 4.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 8.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 132.6 | GO:0005524 | ATP binding(GO:0005524) |
| 0.1 | 43.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 3.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.1 | 2.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 30.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 106.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 6.0 | 286.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 4.6 | 226.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 4.3 | 180.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 3.9 | 74.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 3.8 | 198.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 2.8 | 61.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 1.8 | 70.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 1.6 | 51.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 1.3 | 10.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 1.2 | 236.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 1.1 | 21.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 1.0 | 38.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 1.0 | 29.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 1.0 | 13.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.9 | 13.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.9 | 16.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.9 | 47.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.9 | 33.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.8 | 23.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.8 | 114.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.7 | 83.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.6 | 14.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.5 | 7.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.5 | 14.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.5 | 21.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.5 | 22.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.5 | 39.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.4 | 21.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.4 | 15.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.4 | 20.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.4 | 5.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.3 | 3.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 4.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.3 | 4.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.3 | 11.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.3 | 7.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 10.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.2 | 3.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 5.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 14.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.7 | 691.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 12.0 | 204.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 9.1 | 255.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 7.4 | 154.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 6.7 | 134.2 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 5.7 | 243.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 5.5 | 120.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 4.4 | 74.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 3.8 | 57.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 2.4 | 50.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 1.6 | 28.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 1.5 | 24.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 1.5 | 43.6 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 1.2 | 23.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 1.1 | 37.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 1.0 | 13.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 1.0 | 42.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 1.0 | 23.3 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.9 | 13.0 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.9 | 52.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.8 | 38.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.7 | 17.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.7 | 68.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.6 | 23.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.6 | 20.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.6 | 9.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.5 | 38.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.5 | 8.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.4 | 6.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.4 | 6.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.3 | 5.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.3 | 59.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 3.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 9.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 18.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 5.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 10.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 2.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 10.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 2.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |