Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
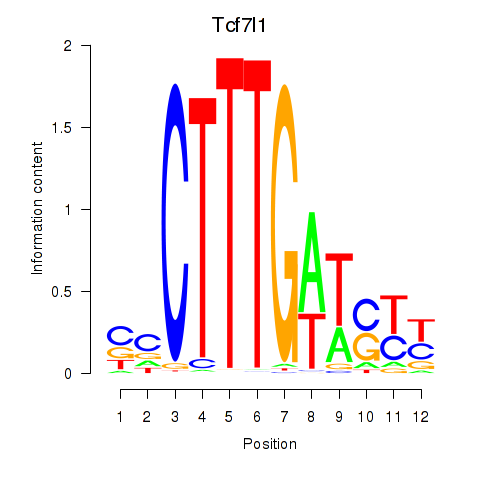
Results for Tcf7l1
Z-value: 0.56
Transcription factors associated with Tcf7l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf7l1
|
ENSRNOG00000014753 | transcription factor 7 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf7l1 | rn6_v1_chr4_-_100660140_100660140 | 0.45 | 1.2e-17 | Click! |
Activity profile of Tcf7l1 motif
Sorted Z-values of Tcf7l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_37645802 | 17.11 |
ENSRNOT00000008022
|
Tcf7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr6_-_99843245 | 11.78 |
ENSRNOT00000080270
|
Gpx2
|
glutathione peroxidase 2 |
| chr9_-_93404883 | 11.09 |
ENSRNOT00000025024
|
Nmur1
|
neuromedin U receptor 1 |
| chr5_-_58987760 | 10.77 |
ENSRNOT00000035040
|
Sit1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr17_+_47870611 | 10.07 |
ENSRNOT00000078555
|
AABR07027872.1
|
|
| chr20_-_31597830 | 9.70 |
ENSRNOT00000085877
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr13_+_83996080 | 9.49 |
ENSRNOT00000004403
ENSRNOT00000070958 |
Cd247
|
Cd247 molecule |
| chrX_-_159891326 | 9.13 |
ENSRNOT00000001154
|
Rbmx
|
RNA binding motif protein, X-linked |
| chr7_-_116201756 | 9.00 |
ENSRNOT00000091401
|
Cyp11b2
|
cytochrome P450, family 11, subfamily b, polypeptide 2 |
| chr20_-_31598118 | 8.89 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr3_-_51297852 | 8.22 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr12_+_24761210 | 8.08 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr4_+_14071507 | 8.05 |
ENSRNOT00000066224
ENSRNOT00000075962 |
Cd36
RGD1565355
|
CD36 molecule similar to fatty acid translocase/CD36 |
| chr15_-_52214616 | 7.98 |
ENSRNOT00000015035
|
Sftpc
|
surfactant protein C |
| chr15_-_41338284 | 7.73 |
ENSRNOT00000077225
|
Tnfrsf19
|
TNF receptor superfamily member 19 |
| chr13_+_53351717 | 7.65 |
ENSRNOT00000012038
|
Kif14
|
kinesin family member 14 |
| chr10_+_31146107 | 7.51 |
ENSRNOT00000008184
|
Adam19
|
ADAM metallopeptidase domain 19 |
| chr3_+_110855000 | 7.42 |
ENSRNOT00000081613
|
Knl1
|
kinetochore scaffold 1 |
| chr4_-_151516894 | 7.02 |
ENSRNOT00000011044
|
Wnt5b
|
wingless-type MMTV integration site family, member 5B |
| chr1_+_101214593 | 6.97 |
ENSRNOT00000028086
|
Tead2
|
TEA domain transcription factor 2 |
| chr4_+_14001761 | 6.82 |
ENSRNOT00000076519
|
Cd36
|
CD36 molecule |
| chr4_+_14151343 | 6.61 |
ENSRNOT00000061687
ENSRNOT00000076573 ENSRNOT00000077219 ENSRNOT00000008319 |
Cd36
|
CD36 molecule |
| chr4_+_78283026 | 6.61 |
ENSRNOT00000059173
|
Gimap8
|
GTPase, IMAP family member 8 |
| chr4_+_14213587 | 6.42 |
ENSRNOT00000067543
|
LOC103690020
|
platelet glycoprotein 4-like |
| chr18_-_15089988 | 6.38 |
ENSRNOT00000074116
|
Mep1b
|
meprin A subunit beta |
| chr3_-_55822551 | 6.12 |
ENSRNOT00000082624
|
Lrp2
|
LDL receptor related protein 2 |
| chr10_+_56453877 | 6.05 |
ENSRNOT00000031640
|
Plscr3
|
phospholipid scramblase 3 |
| chr4_+_92431710 | 5.97 |
ENSRNOT00000049438
|
LOC108350839
|
high mobility group protein B1-like |
| chr5_+_71742911 | 5.93 |
ENSRNOT00000047225
|
Zfp462
|
zinc finger protein 462 |
| chr8_+_22648323 | 5.78 |
ENSRNOT00000013165
|
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr9_+_46657922 | 5.64 |
ENSRNOT00000019180
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr6_-_18351536 | 5.56 |
ENSRNOT00000061588
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr3_+_149741312 | 5.51 |
ENSRNOT00000070854
|
LOC100911637
|
high mobility group protein B1-like |
| chr9_+_46657575 | 5.46 |
ENSRNOT00000083322
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr7_-_132984110 | 5.40 |
ENSRNOT00000046626
|
Hmgb1-ps3
|
high mobility group box 1, pseudogene 3 |
| chr7_+_144628120 | 5.36 |
ENSRNOT00000022247
|
Hoxc5
|
homeo box C5 |
| chr10_+_75365822 | 5.35 |
ENSRNOT00000055705
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chr4_-_77347011 | 5.28 |
ENSRNOT00000008149
|
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr3_+_172856733 | 5.23 |
ENSRNOT00000009826
|
Edn3
|
endothelin 3 |
| chr10_-_109840047 | 5.21 |
ENSRNOT00000054947
|
Notum
|
NOTUM, palmitoleoyl-protein carboxylesterase |
| chr2_-_250235435 | 5.18 |
ENSRNOT00000088618
|
Lmo4
|
LIM domain only 4 |
| chr13_+_5480150 | 5.18 |
ENSRNOT00000057873
|
AABR07019870.1
|
|
| chr1_-_89509343 | 5.14 |
ENSRNOT00000028637
|
Fxyd3
|
FXYD domain-containing ion transport regulator 3 |
| chr7_-_140291620 | 4.96 |
ENSRNOT00000088323
|
Adcy6
|
adenylate cyclase 6 |
| chr1_-_218100272 | 4.86 |
ENSRNOT00000028411
ENSRNOT00000088588 |
Ccnd1
|
cyclin D1 |
| chr16_+_37500017 | 4.85 |
ENSRNOT00000081634
|
AC135696.1
|
high mobility group box 1 (Hmgb1), mRNA |
| chr3_+_149790856 | 4.71 |
ENSRNOT00000075275
|
LOC100911637
|
high mobility group protein B1-like |
| chr17_+_9596957 | 4.66 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr20_+_4039413 | 4.64 |
ENSRNOT00000082136
|
RT1-Bb
|
RT1 class II, locus Bb |
| chr3_+_154043873 | 4.45 |
ENSRNOT00000072502
ENSRNOT00000034166 |
Nnat
|
neuronatin |
| chr4_+_44321883 | 4.44 |
ENSRNOT00000091095
|
Tes
|
testin LIM domain protein |
| chr4_-_60548021 | 4.20 |
ENSRNOT00000065546
|
AABR07060165.1
|
|
| chr10_-_34961349 | 4.18 |
ENSRNOT00000004885
|
LOC103689931
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr7_-_140640953 | 4.18 |
ENSRNOT00000083156
|
Tuba1a
|
tubulin, alpha 1A |
| chr19_-_59985494 | 4.12 |
ENSRNOT00000086591
|
Hmg1l1
|
high-mobility group (nonhistone chromosomal) protein 1-like 1 |
| chr5_-_16799776 | 4.08 |
ENSRNOT00000011721
|
Plag1
|
PLAG1 zinc finger |
| chr2_-_147693082 | 3.95 |
ENSRNOT00000022507
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr5_+_155660553 | 3.92 |
ENSRNOT00000081893
|
Wnt4
|
wingless-type MMTV integration site family, member 4 |
| chr18_-_80865584 | 3.90 |
ENSRNOT00000021750
|
Tshz1
|
teashirt zinc finger homeobox 1 |
| chr16_+_10727571 | 3.76 |
ENSRNOT00000084422
|
Mmrn2
|
multimerin 2 |
| chr10_-_56831243 | 3.73 |
ENSRNOT00000025403
|
Slc16a13
|
solute carrier family 16, member 13 |
| chr3_-_161498951 | 3.73 |
ENSRNOT00000024214
|
Ncoa5
|
nuclear receptor coactivator 5 |
| chr10_-_88172910 | 3.71 |
ENSRNOT00000046956
|
Krt42
|
keratin 42 |
| chr5_-_144479306 | 3.68 |
ENSRNOT00000087697
|
Ago1
|
argonaute 1, RISC catalytic component |
| chr10_+_89358376 | 3.67 |
ENSRNOT00000028067
|
Ifi35
|
interferon-induced protein 35 |
| chr2_+_196013799 | 3.62 |
ENSRNOT00000084023
|
Pogz
|
pogo transposable element with ZNF domain |
| chr5_-_50362344 | 3.61 |
ENSRNOT00000035808
|
Zfp292
|
zinc finger protein 292 |
| chr1_-_170628915 | 3.58 |
ENSRNOT00000042865
|
Dchs1
|
dachsous cadherin-related 1 |
| chr5_-_150167077 | 3.55 |
ENSRNOT00000088493
ENSRNOT00000013761 ENSRNOT00000067349 ENSRNOT00000051097 ENSRNOT00000067189 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr17_-_86657473 | 3.45 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
| chr1_+_141986145 | 3.35 |
ENSRNOT00000030914
|
Sema4b
|
semaphorin 4B |
| chr1_+_187149453 | 3.34 |
ENSRNOT00000082738
|
Xylt1
|
xylosyltransferase 1 |
| chr2_+_142686724 | 3.28 |
ENSRNOT00000014614
|
Proser1
|
proline and serine rich 1 |
| chr8_+_118206698 | 3.22 |
ENSRNOT00000082607
ENSRNOT00000064140 |
Smarcc1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr3_+_21764377 | 3.10 |
ENSRNOT00000063938
|
Gpr21
|
G protein-coupled receptor 21 |
| chr18_+_79773608 | 3.03 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr12_+_9728486 | 3.03 |
ENSRNOT00000001263
|
Lnx2
|
ligand of numb-protein X 2 |
| chr20_-_27208041 | 2.96 |
ENSRNOT00000084720
|
Rufy2
|
RUN and FYVE domain containing 2 |
| chr19_+_39543242 | 2.92 |
ENSRNOT00000018837
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_+_84304228 | 2.90 |
ENSRNOT00000024771
|
Prx
|
periaxin |
| chr6_-_138068648 | 2.85 |
ENSRNOT00000081908
|
Ighm
|
immunoglobulin heavy constant mu |
| chr5_-_147953093 | 2.80 |
ENSRNOT00000075270
|
Khdrbs1
|
KH RNA binding domain containing, signal transduction associated 1 |
| chr5_-_131860637 | 2.78 |
ENSRNOT00000064569
ENSRNOT00000080242 |
Slc5a9
|
solute carrier family 5 member 9 |
| chr8_+_111210811 | 2.76 |
ENSRNOT00000011347
|
Amotl2
|
angiomotin like 2 |
| chr2_+_179952227 | 2.76 |
ENSRNOT00000015081
|
Pdgfc
|
platelet derived growth factor C |
| chr4_+_78240385 | 2.76 |
ENSRNOT00000011041
|
Zfp775
|
zinc finger protein 775 |
| chr1_-_266086299 | 2.76 |
ENSRNOT00000026609
|
Cuedc2
|
CUE domain containing 2 |
| chr1_-_178367828 | 2.73 |
ENSRNOT00000050823
|
AABR07071968.1
|
|
| chr6_-_43493816 | 2.65 |
ENSRNOT00000077786
|
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr2_-_94903226 | 2.63 |
ENSRNOT00000015194
|
AABR07009373.1
|
|
| chr6_-_1622196 | 2.60 |
ENSRNOT00000007492
|
Prkd3
|
protein kinase D3 |
| chr1_-_84008293 | 2.59 |
ENSRNOT00000002053
|
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chrX_-_118513061 | 2.57 |
ENSRNOT00000043338
|
Il13ra2
|
interleukin 13 receptor subunit alpha 2 |
| chr2_+_208738132 | 2.56 |
ENSRNOT00000023972
|
AABR07012795.1
|
|
| chr9_-_116222374 | 2.55 |
ENSRNOT00000090111
ENSRNOT00000067900 |
Arhgap28
|
Rho GTPase activating protein 28 |
| chr9_-_100306194 | 2.52 |
ENSRNOT00000087584
|
RGD1563692
|
similar to hypothetical protein FLJ22671 |
| chr3_+_148541909 | 2.49 |
ENSRNOT00000012187
|
Ccm2l
|
CCM2 like scaffolding protein |
| chr9_-_44126919 | 2.48 |
ENSRNOT00000024701
|
Mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr7_+_12256387 | 2.46 |
ENSRNOT00000021939
|
RGD1359127
|
similar to RIKEN cDNA 2310011J03 |
| chr4_+_57019941 | 2.42 |
ENSRNOT00000011356
|
Smo
|
smoothened, frizzled class receptor |
| chr7_+_144605058 | 2.38 |
ENSRNOT00000031404
|
Hoxc8
|
homeobox C8 |
| chr16_-_74264142 | 2.35 |
ENSRNOT00000026067
|
Dkk4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr13_-_70922245 | 2.33 |
ENSRNOT00000064860
|
Dhx9
|
DExH-box helicase 9 |
| chr10_-_14788617 | 2.32 |
ENSRNOT00000043626
|
Cacna1h
|
calcium voltage-gated channel subunit alpha1 H |
| chr8_-_96132634 | 2.24 |
ENSRNOT00000041655
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chrX_+_156863754 | 2.20 |
ENSRNOT00000083611
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr6_+_108820774 | 2.15 |
ENSRNOT00000089683
ENSRNOT00000080038 ENSRNOT00000006492 |
Ylpm1
|
YLP motif containing 1 |
| chr1_-_134867001 | 2.14 |
ENSRNOT00000090149
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr20_+_48335540 | 2.12 |
ENSRNOT00000000352
|
Cd24
|
CD24 molecule |
| chr11_-_80650802 | 2.07 |
ENSRNOT00000043660
|
Rtp4
|
receptor (chemosensory) transporter protein 4 |
| chr11_-_31213787 | 2.01 |
ENSRNOT00000002800
|
Paxbp1
|
PAX3 and PAX7 binding protein 1 |
| chr7_+_37812831 | 2.00 |
ENSRNOT00000005910
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr8_+_97537968 | 1.97 |
ENSRNOT00000037276
|
Adamts7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7 |
| chr8_+_59457018 | 1.97 |
ENSRNOT00000017900
|
Ireb2
|
iron responsive element binding protein 2 |
| chr8_-_132919110 | 1.95 |
ENSRNOT00000008747
|
Xcr1
|
X-C motif chemokine receptor 1 |
| chr14_-_44845218 | 1.91 |
ENSRNOT00000004003
|
Klhl5
|
kelch-like family member 5 |
| chr7_-_140546908 | 1.90 |
ENSRNOT00000077502
|
Kmt2d
|
lysine methyltransferase 2D |
| chr19_-_55490426 | 1.90 |
ENSRNOT00000081800
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr6_+_137953545 | 1.89 |
ENSRNOT00000006804
|
Crip2
|
cysteine-rich protein 2 |
| chr12_-_11808977 | 1.87 |
ENSRNOT00000071795
|
AABR07035375.1
|
|
| chr16_+_2670618 | 1.81 |
ENSRNOT00000030102
|
Il17rd
|
interleukin 17 receptor D |
| chr15_+_86243148 | 1.80 |
ENSRNOT00000084471
ENSRNOT00000090727 |
Lmo7
|
LIM domain 7 |
| chr1_+_65564173 | 1.78 |
ENSRNOT00000038860
|
Zbtb45
|
zinc finger and BTB domain containing 45 |
| chr10_+_82937971 | 1.76 |
ENSRNOT00000005797
|
Dlx3
|
distal-less homeobox 3 |
| chr1_-_221087449 | 1.76 |
ENSRNOT00000016987
|
Fam89b
|
family with sequence similarity 89, member B |
| chrX_-_128268285 | 1.76 |
ENSRNOT00000009755
ENSRNOT00000081880 |
Thoc2
|
THO complex 2 |
| chr10_-_39405311 | 1.75 |
ENSRNOT00000074616
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr2_-_142686577 | 1.74 |
ENSRNOT00000014562
|
Nhlrc3
|
NHL repeat containing 3 |
| chr4_+_66091641 | 1.74 |
ENSRNOT00000043147
|
AABR07060287.1
|
|
| chr2_+_119112513 | 1.73 |
ENSRNOT00000015460
|
Actl6a
|
actin-like 6A |
| chr13_+_52976507 | 1.69 |
ENSRNOT00000090599
ENSRNOT00000011324 |
Kif21b
|
kinesin family member 21B |
| chr1_-_263431290 | 1.68 |
ENSRNOT00000022633
|
Slc25a28
|
solute carrier family 25 member 28 |
| chr2_-_226779440 | 1.66 |
ENSRNOT00000055669
|
Fnbp1l
|
formin binding protein 1-like |
| chr5_+_173256834 | 1.66 |
ENSRNOT00000089936
|
Ccnl2
|
cyclin L2 |
| chr3_+_80634470 | 1.65 |
ENSRNOT00000045644
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr16_+_75366929 | 1.59 |
ENSRNOT00000049394
|
Defal1
|
defensin alpha-like 1 |
| chr2_+_238257031 | 1.57 |
ENSRNOT00000016066
|
Ints12
|
integrator complex subunit 12 |
| chr3_+_171037957 | 1.56 |
ENSRNOT00000008764
|
Rbm38
|
RNA binding motif protein 38 |
| chr17_-_20364714 | 1.55 |
ENSRNOT00000070962
|
Jarid2
|
jumonji and AT-rich interaction domain containing 2 |
| chr10_-_74413745 | 1.55 |
ENSRNOT00000038296
|
Prr11
|
proline rich 11 |
| chr5_+_116421894 | 1.55 |
ENSRNOT00000080577
ENSRNOT00000086628 ENSRNOT00000004017 |
Nfia
|
nuclear factor I/A |
| chr3_+_3767394 | 1.52 |
ENSRNOT00000067840
|
Gpsm1
|
G-protein signaling modulator 1 |
| chr12_+_25264192 | 1.51 |
ENSRNOT00000079392
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr15_-_82482009 | 1.49 |
ENSRNOT00000011926
|
Dach1
|
dachshund family transcription factor 1 |
| chr1_-_89007041 | 1.48 |
ENSRNOT00000032363
|
Proser3
|
proline and serine rich 3 |
| chr8_+_70630767 | 1.39 |
ENSRNOT00000051353
|
Igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr11_-_78456501 | 1.37 |
ENSRNOT00000036193
|
Tp63
|
tumor protein p63 |
| chrX_-_104984341 | 1.35 |
ENSRNOT00000077996
|
Xkrx
|
XK related, X-linked |
| chr4_+_113968995 | 1.35 |
ENSRNOT00000079511
|
Rtkn
|
rhotekin |
| chr3_+_150188455 | 1.34 |
ENSRNOT00000072917
|
AABR07054370.1
|
|
| chr5_+_128083118 | 1.34 |
ENSRNOT00000079161
|
Zcchc11
|
zinc finger CCHC-type containing 11 |
| chr14_-_51462721 | 1.33 |
ENSRNOT00000015044
|
Hmgb3
|
high mobility group box 3 |
| chr11_+_67465236 | 1.32 |
ENSRNOT00000042374
|
Stfa2
|
stefin A2 |
| chr1_-_198316882 | 1.30 |
ENSRNOT00000085304
ENSRNOT00000064985 |
Taok2
|
TAO kinase 2 |
| chr15_+_105851542 | 1.30 |
ENSRNOT00000086959
|
Rap2a
|
RAS related protein 2a |
| chr2_-_226779266 | 1.28 |
ENSRNOT00000019244
|
Fnbp1l
|
formin binding protein 1-like |
| chr17_-_17947777 | 1.27 |
ENSRNOT00000036876
|
Rnf144b
|
ring finger protein 144B |
| chr6_+_10642700 | 1.26 |
ENSRNOT00000040329
|
LOC100359600
|
karyopherin alpha 2-like |
| chr18_+_30909490 | 1.24 |
ENSRNOT00000026967
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8 |
| chr19_-_41010122 | 1.23 |
ENSRNOT00000088770
|
Vac14
|
Vac14, PIKFYVE complex component |
| chr6_+_80159364 | 1.22 |
ENSRNOT00000005439
|
Pnn
|
pinin, desmosome associated protein |
| chr1_+_221043119 | 1.22 |
ENSRNOT00000028185
|
Map3k11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr3_+_116899878 | 1.21 |
ENSRNOT00000090802
ENSRNOT00000066101 |
Sema6d
|
semaphorin 6D |
| chr20_+_34894419 | 1.21 |
ENSRNOT00000000471
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr7_+_123260607 | 1.19 |
ENSRNOT00000066849
|
Xrcc6
|
X-ray repair cross complementing 6 |
| chr7_-_98098268 | 1.17 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chr9_-_81772851 | 1.17 |
ENSRNOT00000032719
|
Usp37
|
ubiquitin specific peptidase 37 |
| chr2_-_139528162 | 1.16 |
ENSRNOT00000014317
|
Slc7a11
|
solute carrier family 7 member 11 |
| chr13_-_73055631 | 1.15 |
ENSRNOT00000081892
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr12_-_52596076 | 1.15 |
ENSRNOT00000056639
|
Chfr
|
checkpoint with forkhead and ring finger domains |
| chr1_+_128614138 | 1.13 |
ENSRNOT00000076227
ENSRNOT00000078707 |
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr5_-_75676584 | 1.13 |
ENSRNOT00000044348
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr9_+_40817654 | 1.12 |
ENSRNOT00000037392
|
AABR07067339.1
|
|
| chr9_+_66952720 | 1.12 |
ENSRNOT00000023678
|
Nbeal1
|
neurobeachin-like 1 |
| chr2_-_33025271 | 1.11 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_-_72596446 | 1.11 |
ENSRNOT00000008459
|
Hectd1
|
HECT domain E3 ubiquitin protein ligase 1 |
| chrX_-_25628272 | 1.10 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chr12_+_25264400 | 1.07 |
ENSRNOT00000087656
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr4_+_114835064 | 1.04 |
ENSRNOT00000031964
|
LOC103692165
|
rhotekin |
| chr13_-_1946508 | 1.03 |
ENSRNOT00000043890
|
Dsel
|
dermatan sulfate epimerase-like |
| chr12_+_47992091 | 1.02 |
ENSRNOT00000073019
|
Kctd10
|
potassium channel tetramerization domain containing 10 |
| chr11_-_78456200 | 1.00 |
ENSRNOT00000067251
ENSRNOT00000036179 |
Tp63
|
tumor protein p63 |
| chr9_+_94562839 | 0.99 |
ENSRNOT00000022308
|
RGD1311447
|
LOC363276 |
| chr13_-_79801112 | 0.97 |
ENSRNOT00000087323
ENSRNOT00000036483 |
Suco
|
SUN domain containing ossification factor |
| chr15_+_41927241 | 0.96 |
ENSRNOT00000012035
|
Trim13
|
tripartite motif-containing 13 |
| chr13_-_79744939 | 0.96 |
ENSRNOT00000076706
|
Suco
|
SUN domain containing ossification factor |
| chr13_+_89797800 | 0.90 |
ENSRNOT00000005811
|
Usf1
|
upstream transcription factor 1 |
| chr16_+_20027348 | 0.85 |
ENSRNOT00000034589
|
Fam129c
|
family with sequence similarity 129, member C |
| chr6_-_22281886 | 0.82 |
ENSRNOT00000039375
|
Spast
|
spastin |
| chr10_-_87529599 | 0.81 |
ENSRNOT00000074099
|
Krtap2-1
|
keratin associated protein 2-1 |
| chr13_+_99473044 | 0.79 |
ENSRNOT00000004939
|
Cnih4
|
cornichon family AMPA receptor auxiliary protein 4 |
| chr2_-_52282548 | 0.79 |
ENSRNOT00000033627
|
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr3_-_2719135 | 0.77 |
ENSRNOT00000080257
|
Lcn12
|
lipocalin 12 |
| chr1_+_91433030 | 0.76 |
ENSRNOT00000015205
|
Slc7a10
|
solute carrier family 7 member 10 |
| chr20_-_3822754 | 0.75 |
ENSRNOT00000000541
ENSRNOT00000077357 |
Slc39a7
|
solute carrier family 39 member 7 |
| chr5_+_126976456 | 0.73 |
ENSRNOT00000014275
|
Ndc1
|
NDC1 transmembrane nucleoporin |
| chr4_-_77489535 | 0.72 |
ENSRNOT00000008728
|
Pdia4
|
protein disulfide isomerase family A, member 4 |
| chr3_-_2719513 | 0.71 |
ENSRNOT00000020997
|
Lcn12
|
lipocalin 12 |
| chr2_-_186245771 | 0.67 |
ENSRNOT00000066004
ENSRNOT00000079954 |
Dclk2
|
doublecortin-like kinase 2 |
| chr6_-_22281724 | 0.65 |
ENSRNOT00000079137
|
Spast
|
spastin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf7l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 17.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 3.9 | 11.8 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 3.6 | 21.5 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 2.6 | 7.7 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 2.3 | 4.6 | GO:0009608 | response to symbiont(GO:0009608) |
| 1.9 | 5.8 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 1.6 | 8.0 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 1.5 | 9.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 1.3 | 5.2 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 1.3 | 3.9 | GO:0030237 | female sex determination(GO:0030237) |
| 1.2 | 3.6 | GO:0003192 | mitral valve formation(GO:0003192) condensed mesenchymal cell proliferation(GO:0072137) |
| 1.1 | 5.6 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 1.0 | 2.9 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.9 | 2.8 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.9 | 3.7 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.9 | 5.3 | GO:0035983 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.8 | 2.5 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.8 | 4.9 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.8 | 11.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.7 | 2.2 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.7 | 5.0 | GO:1904117 | cellular response to vasopressin(GO:1904117) |
| 0.7 | 2.1 | GO:0002424 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.7 | 4.8 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.7 | 3.9 | GO:0060023 | soft palate development(GO:0060023) |
| 0.6 | 2.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.6 | 1.8 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.6 | 1.8 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.6 | 4.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.6 | 3.9 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.5 | 8.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.5 | 7.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.5 | 1.9 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.5 | 2.8 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.4 | 5.2 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.4 | 2.6 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.4 | 2.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.4 | 3.8 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.4 | 6.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.4 | 2.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.4 | 8.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.4 | 6.1 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.3 | 12.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.3 | 16.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.3 | 1.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.3 | 2.3 | GO:0034650 | aldosterone biosynthetic process(GO:0032342) cortisol metabolic process(GO:0034650) cortisol biosynthetic process(GO:0034651) |
| 0.3 | 3.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.3 | 2.8 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.3 | 3.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.3 | 0.9 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.3 | 12.2 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.3 | 1.2 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.3 | 7.0 | GO:0048339 | hippo signaling(GO:0035329) paraxial mesoderm development(GO:0048339) |
| 0.3 | 5.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.3 | 2.0 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.3 | 2.0 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 | 1.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.3 | 1.9 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.3 | 3.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.3 | 2.6 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.3 | 1.0 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.3 | 0.5 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.2 | 1.5 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 1.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.2 | 4.5 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.2 | 1.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.2 | 1.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 2.6 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.2 | 9.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.2 | 10.8 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.2 | 6.6 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.2 | 6.8 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.2 | 0.7 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.2 | 2.0 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 1.8 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.2 | 0.5 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.2 | 1.7 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 | 1.8 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.2 | 0.6 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.2 | 0.5 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.2 | 0.6 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.2 | 0.3 | GO:0015679 | plasma membrane copper ion transport(GO:0015679) |
| 0.1 | 0.6 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 6.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.2 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 2.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 1.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 1.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 5.9 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 0.8 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 1.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 3.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 2.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 2.1 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.1 | 1.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 1.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.6 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 5.4 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.1 | 6.3 | GO:0001942 | hair follicle development(GO:0001942) skin epidermis development(GO:0098773) |
| 0.1 | 1.9 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 2.4 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 1.7 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 1.7 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 2.9 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 3.0 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 1.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 1.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 2.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 1.0 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 2.7 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 4.2 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 1.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 1.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 1.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 3.7 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 1.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 2.2 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 1.9 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 2.0 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 1.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.8 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 1.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.5 | GO:0097502 | mannosylation(GO:0097502) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 1.8 | 9.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 1.2 | 9.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.0 | 5.8 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.6 | 17.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.6 | 4.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.6 | 8.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.5 | 3.7 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.5 | 5.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.4 | 7.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.4 | 6.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 8.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 2.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 6.8 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.3 | 27.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 1.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.3 | 4.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 7.7 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 0.6 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.2 | 2.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 14.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 1.8 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 6.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.9 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.8 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.5 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 1.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.9 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 6.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 2.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 8.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 3.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 7.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 17.4 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 14.1 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 3.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 6.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 4.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 3.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 7.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 3.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 18.8 | GO:0005654 | nucleoplasm(GO:0005654) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 21.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 3.0 | 9.0 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 1.7 | 5.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.4 | 10.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.1 | 3.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 1.0 | 3.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 1.0 | 6.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.8 | 5.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.8 | 6.4 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.7 | 2.2 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.6 | 2.6 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.5 | 5.3 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.5 | 11.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.5 | 2.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.5 | 7.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.5 | 2.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.4 | 1.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.4 | 2.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.4 | 1.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.4 | 7.0 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.4 | 5.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.4 | 4.6 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.3 | 1.0 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 1.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 2.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.3 | 1.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.3 | 1.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 2.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.3 | 6.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 8.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 2.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.3 | 10.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 10.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 3.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 11.1 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.2 | 6.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 0.6 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.2 | 1.7 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 1.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 1.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 2.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 5.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 28.2 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 2.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.8 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 8.7 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 14.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 18.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 2.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 4.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 3.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 1.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 1.9 | GO:0004950 | chemokine receptor activity(GO:0004950) |
| 0.1 | 1.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 3.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.8 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 2.8 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 1.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 2.5 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 1.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 0.7 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 2.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 2.5 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 3.8 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 2.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 5.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 8.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.8 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 4.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 8.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 5.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 2.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.3 | 14.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 9.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 16.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 5.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 11.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 20.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 5.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 4.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 5.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 15.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 10.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 2.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 3.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 9.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.4 | 10.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 5.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.3 | 21.5 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.2 | 8.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 14.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 6.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 2.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 4.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 2.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 14.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 18.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 6.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.8 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 1.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.1 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |