Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
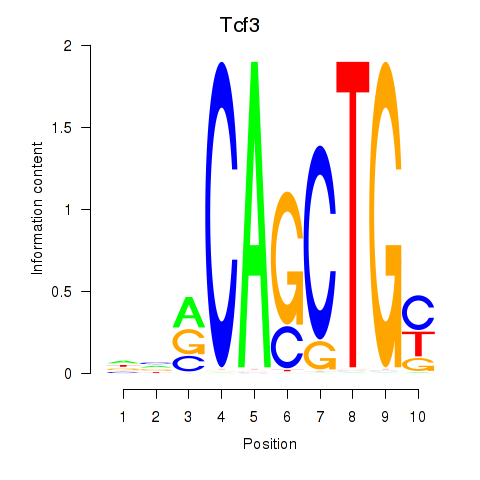
Results for Tcf3
Z-value: 1.19
Transcription factors associated with Tcf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf3
|
ENSRNOG00000051499 | transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf3 | rn6_v1_chr7_+_12144162_12144162 | 0.23 | 3.3e-05 | Click! |
Activity profile of Tcf3 motif
Sorted Z-values of Tcf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_59025631 | 76.48 |
ENSRNOT00000049000
ENSRNOT00000022801 |
Tpm2
|
tropomyosin 2, beta |
| chr1_+_80321585 | 58.33 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr4_+_144382945 | 51.04 |
ENSRNOT00000007601
|
Cav3
|
caveolin 3 |
| chr9_+_82556573 | 48.24 |
ENSRNOT00000026860
|
Des
|
desmin |
| chr9_+_14529218 | 43.86 |
ENSRNOT00000016893
|
Apobec2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| chr6_-_26624092 | 42.83 |
ENSRNOT00000008113
|
Trim54
|
tripartite motif-containing 54 |
| chr10_-_95934345 | 41.86 |
ENSRNOT00000004349
|
Cacng1
|
calcium voltage-gated channel auxiliary subunit gamma 1 |
| chr1_-_254735548 | 40.55 |
ENSRNOT00000025258
|
Ankrd1
|
ankyrin repeat domain 1 |
| chr4_-_129619142 | 40.52 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chrX_+_71342775 | 39.97 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr16_-_49574314 | 36.91 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr13_+_52889737 | 36.53 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr9_-_82699551 | 34.48 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chr15_+_34251606 | 33.84 |
ENSRNOT00000025725
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr1_+_222311253 | 29.97 |
ENSRNOT00000028749
|
Macrod1
|
MACRO domain containing 1 |
| chr3_+_79860179 | 29.95 |
ENSRNOT00000081160
ENSRNOT00000068444 |
Rapsn
|
receptor-associated protein of the synapse |
| chr3_-_161246351 | 29.62 |
ENSRNOT00000020348
|
Tnnc2
|
troponin C2, fast skeletal type |
| chr19_-_55510460 | 29.58 |
ENSRNOT00000019820
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr13_-_84331905 | 29.15 |
ENSRNOT00000004965
|
Dusp27
|
dual specificity phosphatase 27 (putative) |
| chr10_-_94557764 | 28.49 |
ENSRNOT00000016841
|
Scn4a
|
sodium voltage-gated channel alpha subunit 4 |
| chr3_-_117766120 | 27.37 |
ENSRNOT00000056022
|
Fbn1
|
fibrillin 1 |
| chr11_+_66713888 | 27.34 |
ENSRNOT00000003340
|
Fbxo40
|
F-box protein 40 |
| chr10_-_8498422 | 27.25 |
ENSRNOT00000082332
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr1_+_167197549 | 26.11 |
ENSRNOT00000027427
|
Art1
|
ADP-ribosyltransferase 1 |
| chr8_+_130416355 | 25.76 |
ENSRNOT00000026234
|
Klhl40
|
kelch-like family member 40 |
| chr1_+_215609645 | 25.23 |
ENSRNOT00000076140
ENSRNOT00000027487 |
Tnni2
|
troponin I2, fast skeletal type |
| chr1_+_222310920 | 25.10 |
ENSRNOT00000091465
|
Macrod1
|
MACRO domain containing 1 |
| chrX_-_111942749 | 24.50 |
ENSRNOT00000087583
|
AABR07040840.1
|
|
| chr1_+_215609036 | 24.27 |
ENSRNOT00000076187
|
Tnni2
|
troponin I2, fast skeletal type |
| chr11_+_27208564 | 24.25 |
ENSRNOT00000002158
|
Map3k7cl
|
MAP3K7 C-terminal like |
| chr13_+_52662996 | 23.76 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr9_-_115850634 | 23.56 |
ENSRNOT00000049720
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr7_+_120153184 | 22.86 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chrX_-_72515320 | 22.61 |
ENSRNOT00000076725
|
Phka1
|
phosphorylase kinase, alpha 1 |
| chr3_-_148407778 | 22.51 |
ENSRNOT00000011262
|
Foxs1
|
forkhead box S1 |
| chr10_+_92628356 | 22.16 |
ENSRNOT00000072480
|
Myl4
|
myosin, light chain 4 |
| chr2_+_11658568 | 21.65 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr10_+_36098051 | 21.56 |
ENSRNOT00000083971
|
Adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr6_-_99783047 | 21.49 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr3_-_72219246 | 21.20 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chrX_-_72515057 | 21.06 |
ENSRNOT00000076850
ENSRNOT00000041138 |
Phka1
|
phosphorylase kinase, alpha 1 |
| chr10_-_89338739 | 20.83 |
ENSRNOT00000073923
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr10_-_8654892 | 20.67 |
ENSRNOT00000066534
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr15_+_4240203 | 20.08 |
ENSRNOT00000010178
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr8_-_21831668 | 19.98 |
ENSRNOT00000027897
|
Col5a3
|
collagen type V alpha 3 chain |
| chr10_+_34185898 | 19.75 |
ENSRNOT00000003339
|
Trim7
|
tripartite motif-containing 7 |
| chr1_+_220446425 | 19.25 |
ENSRNOT00000027339
|
LOC100911932
|
endosialin-like |
| chr5_+_48224994 | 18.94 |
ENSRNOT00000068028
|
Rragd
|
Ras-related GTP binding D |
| chr8_-_84632817 | 18.66 |
ENSRNOT00000076942
|
Mlip
|
muscular LMNA-interacting protein |
| chr6_-_125723944 | 18.59 |
ENSRNOT00000007004
|
Fbln5
|
fibulin 5 |
| chr16_+_6962722 | 18.49 |
ENSRNOT00000023330
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr6_-_125723732 | 18.16 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr2_-_88135410 | 17.75 |
ENSRNOT00000014180
|
Car3
|
carbonic anhydrase 3 |
| chr2_+_178679395 | 17.53 |
ENSRNOT00000088686
|
Fam198b
|
family with sequence similarity 198, member B |
| chr1_-_215846911 | 17.45 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr1_-_265298797 | 17.06 |
ENSRNOT00000023137
|
Poll
|
DNA polymerase lambda |
| chr4_+_57952982 | 16.86 |
ENSRNOT00000014465
|
Cpa1
|
carboxypeptidase A1 |
| chr18_-_31430973 | 16.68 |
ENSRNOT00000026065
|
Pcdh12
|
protocadherin 12 |
| chr8_-_62948720 | 16.46 |
ENSRNOT00000075476
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr1_+_220353356 | 16.36 |
ENSRNOT00000080189
|
Cd248
|
CD248 molecule |
| chr13_+_85818427 | 16.30 |
ENSRNOT00000077227
ENSRNOT00000006117 |
Rxrg
|
retinoid X receptor gamma |
| chr1_+_266952561 | 16.27 |
ENSRNOT00000076452
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr4_-_82202096 | 15.97 |
ENSRNOT00000081824
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr8_+_22368745 | 15.23 |
ENSRNOT00000049973
|
Slc44a2
|
solute carrier family 44 member 2 |
| chr20_+_3553455 | 14.88 |
ENSRNOT00000090080
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_-_172361779 | 14.77 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr1_+_220362064 | 14.59 |
ENSRNOT00000074361
|
LOC108348067
|
endosialin |
| chr8_+_39997875 | 14.35 |
ENSRNOT00000050609
|
Esam
|
endothelial cell adhesion molecule |
| chr1_-_89559960 | 14.14 |
ENSRNOT00000092133
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr19_-_43911057 | 13.99 |
ENSRNOT00000026017
|
Ctrb1
|
chymotrypsinogen B1 |
| chr10_-_89816491 | 13.80 |
ENSRNOT00000028220
|
Meox1
|
mesenchyme homeobox 1 |
| chr8_-_49025917 | 13.75 |
ENSRNOT00000078816
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr3_+_5624506 | 13.64 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr10_-_18063391 | 13.48 |
ENSRNOT00000072297
|
Fgf18
|
fibroblast growth factor 18 |
| chr5_-_48436531 | 13.47 |
ENSRNOT00000060914
|
Pm20d2
|
peptidase M20 domain containing 2 |
| chrX_+_63520991 | 13.46 |
ENSRNOT00000071590
|
Apoo
|
apolipoprotein O |
| chr1_+_101884276 | 13.31 |
ENSRNOT00000082917
|
Tmem143
|
transmembrane protein 143 |
| chr6_+_6427657 | 13.17 |
ENSRNOT00000005664
|
Pkdcc
|
protein kinase domain containing, cytoplasmic |
| chr1_-_128695995 | 13.17 |
ENSRNOT00000077020
|
Synm
|
synemin |
| chr11_+_3119885 | 13.16 |
ENSRNOT00000042316
|
Vgll3
|
vestigial-like family member 3 |
| chr10_+_54126486 | 13.16 |
ENSRNOT00000078820
|
Gas7
|
growth arrest specific 7 |
| chr3_-_80933283 | 13.14 |
ENSRNOT00000007771
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr3_-_110556808 | 13.11 |
ENSRNOT00000092158
ENSRNOT00000045362 |
RGD1565536
|
similar to hypothetical protein |
| chr6_-_86822094 | 12.81 |
ENSRNOT00000006531
|
Fkbp3
|
FK506 binding protein 3 |
| chr3_-_7141522 | 12.75 |
ENSRNOT00000014572
|
Cel
|
carboxyl ester lipase |
| chr4_+_70689737 | 12.61 |
ENSRNOT00000018852
|
Prss2
|
protease, serine, 2 |
| chr18_+_25163561 | 12.59 |
ENSRNOT00000042287
ENSRNOT00000049898 ENSRNOT00000017573 |
Bin1
|
bridging integrator 1 |
| chr15_+_39945095 | 12.41 |
ENSRNOT00000016826
|
Shisa2
|
shisa family member 2 |
| chr5_+_169181418 | 12.30 |
ENSRNOT00000004508
|
Klhl21
|
kelch-like family member 21 |
| chr13_-_91020112 | 12.30 |
ENSRNOT00000030449
|
Dusp23
|
dual specificity phosphatase 23 |
| chr6_+_98284170 | 12.26 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr9_+_111028824 | 11.99 |
ENSRNOT00000041418
ENSRNOT00000056457 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr4_-_160605107 | 11.68 |
ENSRNOT00000090725
|
Tspan9
|
tetraspanin 9 |
| chrX_-_159841072 | 11.65 |
ENSRNOT00000001158
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr2_-_231521052 | 11.61 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chrX_+_53360839 | 11.38 |
ENSRNOT00000091467
ENSRNOT00000034372 ENSRNOT00000081061 |
Dmd
|
dystrophin |
| chr4_-_14490446 | 11.36 |
ENSRNOT00000009132
|
Sema3c
|
semaphorin 3C |
| chr9_+_111028575 | 11.28 |
ENSRNOT00000043451
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr14_+_60764409 | 11.19 |
ENSRNOT00000005168
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr16_-_71319449 | 11.10 |
ENSRNOT00000029284
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr16_-_71319052 | 11.00 |
ENSRNOT00000050980
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr13_-_111581018 | 10.89 |
ENSRNOT00000083072
ENSRNOT00000077981 |
Sertad4
|
SERTA domain containing 4 |
| chr11_+_46009322 | 10.84 |
ENSRNOT00000002233
|
Tmem45a
|
transmembrane protein 45A |
| chr4_-_38240848 | 10.82 |
ENSRNOT00000007567
|
Ndufa4
|
NADH:ubiquinone oxidoreductase subunit A4 |
| chr16_-_59721227 | 10.78 |
ENSRNOT00000015166
|
Prag1
|
PEAK1 related kinase activating pseudokinase 1 |
| chr9_+_82700468 | 10.65 |
ENSRNOT00000027227
|
Inha
|
inhibin alpha subunit |
| chr11_+_46714355 | 10.47 |
ENSRNOT00000073404
|
Tmem45al
|
transmembrane protein 45A-like |
| chr7_+_123043503 | 10.43 |
ENSRNOT00000026258
ENSRNOT00000086355 |
Tef
|
TEF, PAR bZIP transcription factor |
| chrX_-_25628272 | 10.28 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chr20_+_18780605 | 10.28 |
ENSRNOT00000000754
|
Bicc1
|
BicC family RNA binding protein 1 |
| chr2_-_105089659 | 10.15 |
ENSRNOT00000043381
|
Cpb1
|
carboxypeptidase B1 |
| chr3_-_9037942 | 10.07 |
ENSRNOT00000036770
|
Ier5l
|
immediate early response 5-like |
| chr5_-_79874671 | 9.94 |
ENSRNOT00000084563
|
Tnc
|
tenascin C |
| chr15_+_37806836 | 9.72 |
ENSRNOT00000076285
|
Il17d
|
interleukin 17D |
| chr10_-_110101872 | 9.72 |
ENSRNOT00000071893
|
Ccdc57
|
coiled-coil domain containing 57 |
| chr3_-_5617225 | 9.65 |
ENSRNOT00000008327
|
Tmem8c
|
transmembrane protein 8C |
| chr20_+_40236437 | 9.53 |
ENSRNOT00000000743
|
Hs3st5
|
heparan sulfate-glucosamine 3-sulfotransferase 5 |
| chr5_+_75392790 | 9.45 |
ENSRNOT00000048457
|
Musk
|
muscle associated receptor tyrosine kinase |
| chr8_-_116993193 | 9.41 |
ENSRNOT00000026327
|
Dag1
|
dystroglycan 1 |
| chr9_+_42871950 | 9.34 |
ENSRNOT00000089673
|
Arid5a
|
AT-rich interaction domain 5A |
| chr7_+_122157201 | 9.31 |
ENSRNOT00000025193
|
Adsl
|
adenylosuccinate lyase |
| chr7_-_139223116 | 9.21 |
ENSRNOT00000086559
|
Endou
|
endonuclease, poly(U)-specific |
| chr19_-_56772904 | 9.08 |
ENSRNOT00000024232
|
Abcb10
|
ATP binding cassette subfamily B member 10 |
| chr1_-_277867630 | 9.03 |
ENSRNOT00000054679
ENSRNOT00000091038 ENSRNOT00000079869 |
Ablim1
|
actin-binding LIM protein 1 |
| chr5_-_144274981 | 8.98 |
ENSRNOT00000065871
|
Map7d1
|
MAP7 domain containing 1 |
| chr4_-_147163467 | 8.82 |
ENSRNOT00000010748
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr5_+_147476221 | 8.78 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr10_-_82197848 | 8.72 |
ENSRNOT00000081307
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr18_+_52215682 | 8.69 |
ENSRNOT00000037901
|
Megf10
|
multiple EGF-like domains 10 |
| chr9_+_82571269 | 8.58 |
ENSRNOT00000026941
|
Speg
|
SPEG complex locus |
| chr8_-_22874637 | 8.55 |
ENSRNOT00000064551
ENSRNOT00000090424 |
Dock6
|
dedicator of cytokinesis 6 |
| chr18_+_24708115 | 8.51 |
ENSRNOT00000061054
|
Lims2
|
LIM zinc finger domain containing 2 |
| chr10_+_104451222 | 8.48 |
ENSRNOT00000088665
ENSRNOT00000071694 |
Smim5
|
small integral membrane protein 5 |
| chr3_+_11207542 | 8.41 |
ENSRNOT00000013546
|
Prrc2b
|
proline-rich coiled-coil 2B |
| chr10_+_94566928 | 8.36 |
ENSRNOT00000078446
|
Prr29
|
proline rich 29 |
| chr4_+_129574264 | 8.33 |
ENSRNOT00000010185
|
Arl6ip5
|
ADP-ribosylation factor like GTPase 6 interacting protein 5 |
| chr1_-_260638816 | 8.24 |
ENSRNOT00000065632
ENSRNOT00000017938 ENSRNOT00000077962 |
Pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr14_+_13192347 | 8.12 |
ENSRNOT00000000092
|
Antxr2
|
anthrax toxin receptor 2 |
| chr2_-_257376756 | 8.02 |
ENSRNOT00000065811
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr8_-_53146953 | 8.00 |
ENSRNOT00000045356
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr12_+_24473981 | 7.97 |
ENSRNOT00000001973
|
Fzd9
|
frizzled class receptor 9 |
| chr4_-_159697207 | 7.95 |
ENSRNOT00000086440
|
Ccnd2
|
cyclin D2 |
| chr1_-_277768368 | 7.93 |
ENSRNOT00000023113
|
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr13_-_35048444 | 7.77 |
ENSRNOT00000009963
|
Gli2
|
GLI family zinc finger 2 |
| chr14_-_104523297 | 7.73 |
ENSRNOT00000039523
|
Cep68
|
centrosomal protein 68 |
| chr4_-_70747226 | 7.72 |
ENSRNOT00000044960
|
LOC102554637
|
anionic trypsin-2-like |
| chr6_-_21135880 | 7.54 |
ENSRNOT00000051239
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chrX_-_16929907 | 7.52 |
ENSRNOT00000003971
|
Shroom4
|
shroom family member 4 |
| chr4_-_82215022 | 7.49 |
ENSRNOT00000010256
|
Hoxa11
|
homeobox A11 |
| chr16_-_81912738 | 7.49 |
ENSRNOT00000087095
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr7_+_59349745 | 7.43 |
ENSRNOT00000085334
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr5_+_156215417 | 7.37 |
ENSRNOT00000067616
ENSRNOT00000077742 |
Ece1
|
endothelin converting enzyme 1 |
| chr12_-_38995570 | 7.35 |
ENSRNOT00000001806
|
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chr4_+_83713666 | 7.31 |
ENSRNOT00000086473
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr1_-_19376301 | 7.29 |
ENSRNOT00000015547
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr1_-_214971929 | 7.28 |
ENSRNOT00000027195
|
Mob2
|
MOB kinase activator 2 |
| chr10_+_109446444 | 7.11 |
ENSRNOT00000066736
|
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr8_-_22974321 | 7.05 |
ENSRNOT00000017369
|
Epor
|
erythropoietin receptor |
| chr20_-_49486550 | 7.05 |
ENSRNOT00000048270
ENSRNOT00000076541 |
Prdm1
|
PR/SET domain 1 |
| chr1_+_72420352 | 7.04 |
ENSRNOT00000066307
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr15_+_23792931 | 7.02 |
ENSRNOT00000092091
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr20_-_4863198 | 7.01 |
ENSRNOT00000001108
|
Ltb
|
lymphotoxin beta |
| chr9_+_84689150 | 6.98 |
ENSRNOT00000072242
|
Kcne4
|
potassium voltage-gated channel subfamily E regulatory subunit 4 |
| chr17_+_44794130 | 6.88 |
ENSRNOT00000077571
|
Hist1h2ac
|
histone cluster 1, H2ac |
| chr3_-_60166013 | 6.79 |
ENSRNOT00000024922
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr5_-_164959851 | 6.66 |
ENSRNOT00000012301
|
Fbxo6
|
F-box protein 6 |
| chr18_+_79813759 | 6.60 |
ENSRNOT00000021768
|
Zfp516
|
zinc finger protein 516 |
| chr20_-_4863011 | 6.56 |
ENSRNOT00000079503
|
Ltb
|
lymphotoxin beta |
| chr1_+_101214593 | 6.42 |
ENSRNOT00000028086
|
Tead2
|
TEA domain transcription factor 2 |
| chr9_-_82419288 | 6.41 |
ENSRNOT00000004797
|
Tuba4a
|
tubulin, alpha 4A |
| chr6_+_35314444 | 6.41 |
ENSRNOT00000005666
|
Osr1
|
odd-skipped related transciption factor 1 |
| chr3_+_58084606 | 6.38 |
ENSRNOT00000084797
|
Metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr2_+_198417619 | 6.15 |
ENSRNOT00000085945
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr2_+_198390166 | 6.14 |
ENSRNOT00000081042
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr19_+_52032886 | 6.03 |
ENSRNOT00000019923
|
Mlycd
|
malonyl-CoA decarboxylase |
| chrX_-_111191932 | 6.01 |
ENSRNOT00000088050
ENSRNOT00000083613 |
Morc4
|
MORC family CW-type zinc finger 4 |
| chr1_+_78546012 | 6.01 |
ENSRNOT00000058172
|
Gng5
|
G protein subunit gamma 5 |
| chrX_-_134866210 | 5.95 |
ENSRNOT00000005331
|
Apln
|
apelin |
| chr13_+_52147555 | 5.85 |
ENSRNOT00000084766
|
Lmod1
|
leiomodin 1 |
| chr16_-_19918644 | 5.82 |
ENSRNOT00000083345
ENSRNOT00000023926 |
Plvap
|
plasmalemma vesicle associated protein |
| chr15_-_61772516 | 5.70 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chr4_+_71795768 | 5.64 |
ENSRNOT00000031921
|
Tas2r135
|
taste receptor, type 2, member 135 |
| chr5_+_18901039 | 5.63 |
ENSRNOT00000012066
|
Fam110b
|
family with sequence similarity 110, member B |
| chr10_-_90312386 | 5.62 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr10_+_104359342 | 5.55 |
ENSRNOT00000006327
|
Tsen54
|
tRNA splicing endonuclease subunit 54 |
| chr10_+_90930010 | 5.51 |
ENSRNOT00000003765
|
Higd1b
|
HIG1 hypoxia inducible domain family, member 1B |
| chr2_-_174413038 | 5.47 |
ENSRNOT00000089229
ENSRNOT00000037115 |
Golim4
|
golgi integral membrane protein 4 |
| chr10_+_59173268 | 5.45 |
ENSRNOT00000013486
|
Ube2g1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr7_+_3332788 | 5.42 |
ENSRNOT00000010180
|
Cd63
|
Cd63 molecule |
| chr12_-_25638797 | 5.29 |
ENSRNOT00000002033
|
Gatsl2
|
GATS protein-like 2 |
| chr4_+_181481147 | 5.27 |
ENSRNOT00000002523
|
Klhl42
|
kelch-like family, member 42 |
| chrX_-_70233073 | 5.27 |
ENSRNOT00000004048
|
Awat2
|
acyl-CoA wax alcohol acyltransferase 2 |
| chr19_-_37907714 | 5.21 |
ENSRNOT00000026361
|
Ctrl
|
chymotrypsin-like |
| chr1_+_128924966 | 5.18 |
ENSRNOT00000019267
|
Igf1r
|
insulin-like growth factor 1 receptor |
| chr4_-_19413484 | 5.15 |
ENSRNOT00000009489
|
Sema3d
|
semaphorin 3D |
| chr14_+_36047144 | 5.04 |
ENSRNOT00000003088
|
Lnx1
|
ligand of numb-protein X 1 |
| chr4_-_70628470 | 4.98 |
ENSRNOT00000029319
|
Try5
|
trypsin 5 |
| chr2_+_201172312 | 4.91 |
ENSRNOT00000026458
|
Wars2
|
tryptophanyl tRNA synthetase 2 (mitochondrial) |
| chr10_-_88307412 | 4.90 |
ENSRNOT00000089933
|
Jup
|
junction plakoglobin |
| chr10_-_88307132 | 4.90 |
ENSRNOT00000040845
|
Jup
|
junction plakoglobin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.8 | 55.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 12.8 | 51.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 11.0 | 43.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 9.1 | 27.4 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 7.9 | 23.8 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 7.4 | 22.1 | GO:0035607 | ventricular zone neuroblast division(GO:0021847) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) |
| 5.8 | 23.3 | GO:0031179 | peptide modification(GO:0031179) |
| 5.8 | 46.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 5.7 | 11.4 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 5.4 | 21.6 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 5.2 | 36.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 4.9 | 34.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 4.8 | 33.8 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 4.6 | 18.5 | GO:1902730 | positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 4.6 | 22.9 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 4.5 | 13.5 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 4.4 | 43.7 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 4.3 | 4.3 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 4.2 | 12.6 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 3.8 | 19.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 3.8 | 11.4 | GO:0016203 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 3.7 | 51.9 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 3.7 | 36.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 3.6 | 28.5 | GO:0015871 | choline transport(GO:0015871) |
| 3.5 | 7.1 | GO:1904373 | response to kainic acid(GO:1904373) |
| 3.3 | 9.9 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 3.3 | 29.6 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 3.2 | 9.6 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 3.2 | 6.4 | GO:0036023 | embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 3.2 | 9.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 3.2 | 18.9 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 3.1 | 9.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 2.9 | 11.6 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.8 | 14.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 2.8 | 13.8 | GO:0061056 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 2.7 | 47.9 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 2.6 | 13.2 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 2.6 | 7.8 | GO:0021508 | floor plate formation(GO:0021508) |
| 2.5 | 47.9 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 2.5 | 17.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 2.3 | 25.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 2.2 | 13.2 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 2.2 | 13.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 2.2 | 8.7 | GO:0014816 | skeletal muscle satellite cell activation(GO:0014719) skeletal muscle satellite cell differentiation(GO:0014816) |
| 2.1 | 27.9 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 2.1 | 6.4 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 2.1 | 8.3 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 2.0 | 6.0 | GO:0033869 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) fatty-acyl-CoA catabolic process(GO:0036115) |
| 2.0 | 8.0 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 2.0 | 9.8 | GO:0002159 | desmosome assembly(GO:0002159) |
| 1.9 | 48.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 1.9 | 74.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 1.7 | 5.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 1.7 | 13.6 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 1.7 | 8.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 1.6 | 4.9 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 1.6 | 41.8 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 1.6 | 98.6 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 1.5 | 4.6 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 1.5 | 10.7 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 1.5 | 6.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 1.5 | 5.9 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 1.5 | 8.7 | GO:0010045 | response to nickel cation(GO:0010045) |
| 1.4 | 49.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 1.4 | 5.6 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 1.3 | 3.9 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 1.2 | 2.5 | GO:1904412 | canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cardiac ventricle development(GO:1904412) |
| 1.2 | 3.7 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 1.2 | 15.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 1.2 | 8.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 1.1 | 5.6 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 1.1 | 8.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 1.1 | 58.3 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 1.0 | 3.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 1.0 | 12.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 1.0 | 7.0 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) |
| 1.0 | 17.1 | GO:0048535 | lymph node development(GO:0048535) |
| 1.0 | 26.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 1.0 | 12.8 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 1.0 | 10.7 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 1.0 | 2.9 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 1.0 | 4.8 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.9 | 16.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.9 | 12.8 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.8 | 4.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.8 | 13.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.8 | 4.8 | GO:2000504 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.8 | 14.8 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.8 | 17.9 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.8 | 1.5 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.8 | 4.5 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) |
| 0.8 | 3.8 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.8 | 6.8 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.7 | 17.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.7 | 21.5 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.7 | 7.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.7 | 5.4 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.6 | 1.9 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.6 | 16.3 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.6 | 4.9 | GO:0034351 | negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.6 | 8.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.6 | 9.0 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.6 | 7.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.6 | 2.9 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.6 | 4.6 | GO:0061193 | taste bud development(GO:0061193) |
| 0.5 | 21.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.5 | 22.5 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.5 | 2.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.5 | 2.5 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.5 | 6.4 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.5 | 1.5 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.5 | 4.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.5 | 21.2 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.5 | 29.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.5 | 12.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 1.8 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.4 | 6.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.4 | 5.8 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.4 | 8.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.4 | 10.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.4 | 5.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.4 | 12.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.4 | 8.8 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.3 | 2.8 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.3 | 7.9 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.3 | 2.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.3 | 1.0 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.3 | 13.9 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.3 | 14.9 | GO:0032094 | response to food(GO:0032094) |
| 0.3 | 19.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.3 | 1.7 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.3 | 7.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 35.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.3 | 5.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.3 | 11.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 2.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 4.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.2 | 1.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 1.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 2.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 6.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 1.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 3.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 18.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.2 | 3.6 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.2 | 1.5 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.2 | 8.6 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.2 | 0.9 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 3.1 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.2 | 4.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 0.6 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.2 | 19.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.2 | 0.5 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 1.5 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) |
| 0.2 | 16.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 9.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 2.5 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.1 | 1.1 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 5.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 1.0 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 1.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 0.6 | GO:0031033 | myosin filament organization(GO:0031033) myosin filament assembly(GO:0031034) |
| 0.1 | 3.0 | GO:0042462 | gamma-aminobutyric acid signaling pathway(GO:0007214) eye photoreceptor cell development(GO:0042462) |
| 0.1 | 19.2 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.1 | 10.8 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 1.6 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 1.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 3.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 8.2 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.1 | 4.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 1.8 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 2.7 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 7.6 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 3.6 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 3.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 3.1 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.8 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.1 | GO:0031627 | telomeric loop formation(GO:0031627) regulation of DNA strand elongation(GO:0060382) positive regulation of DNA strand elongation(GO:0060383) |
| 0.0 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 1.4 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.9 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.1 | GO:0051198 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 0.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.9 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.4 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 76.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 9.6 | 48.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 8.6 | 102.9 | GO:0005861 | troponin complex(GO:0005861) |
| 7.4 | 36.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 5.7 | 34.5 | GO:1990393 | 3M complex(GO:1990393) |
| 5.5 | 43.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 5.0 | 20.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 3.8 | 71.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 3.6 | 10.7 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin complex(GO:0043511) |
| 3.1 | 21.5 | GO:0008091 | spectrin(GO:0008091) |
| 2.6 | 13.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 2.5 | 27.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 2.5 | 9.8 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 2.2 | 121.2 | GO:0031672 | A band(GO:0031672) |
| 2.1 | 6.4 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 2.1 | 42.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 2.0 | 90.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 2.0 | 6.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.9 | 13.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.8 | 12.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 1.5 | 4.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 1.5 | 20.0 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 1.5 | 172.4 | GO:0031674 | I band(GO:0031674) |
| 1.4 | 5.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.3 | 36.3 | GO:0030017 | sarcomere(GO:0030017) |
| 1.2 | 7.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.2 | 4.8 | GO:0031592 | centrosomal corona(GO:0031592) |
| 1.1 | 13.8 | GO:0045180 | basal cortex(GO:0045180) |
| 1.1 | 5.6 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 1.1 | 7.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 1.0 | 5.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 1.0 | 1.9 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.8 | 6.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.7 | 3.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.7 | 10.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.6 | 2.5 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.6 | 7.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.6 | 9.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.5 | 7.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.5 | 4.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.4 | 12.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.4 | 12.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.4 | 2.8 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.4 | 1.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.3 | 10.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.3 | 2.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 2.4 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.3 | 2.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 20.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 8.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 4.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.3 | 53.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 1.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 1.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 14.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 1.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 2.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 1.5 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 1.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 4.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 19.0 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 12.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 14.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 21.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.2 | 9.1 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.2 | 7.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 9.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 15.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 8.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 3.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 5.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 7.3 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 136.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 30.1 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.1 | 10.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 16.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 3.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 4.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 13.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.9 | 23.8 | GO:0030172 | troponin C binding(GO:0030172) |
| 8.7 | 43.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 8.3 | 58.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 8.1 | 32.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 7.3 | 43.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 7.1 | 49.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 6.4 | 51.0 | GO:0071253 | connexin binding(GO:0071253) |
| 4.8 | 67.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 4.4 | 8.7 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 4.3 | 12.8 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 4.1 | 40.5 | GO:0031432 | titin binding(GO:0031432) |
| 4.0 | 181.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 3.8 | 22.9 | GO:0048030 | disaccharide binding(GO:0048030) |
| 3.3 | 29.9 | GO:0043495 | protein anchor(GO:0043495) |
| 3.3 | 26.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 2.8 | 14.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 2.8 | 22.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 2.7 | 16.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 2.7 | 13.6 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 2.7 | 13.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 2.7 | 18.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 2.4 | 17.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 2.2 | 17.7 | GO:0016151 | nickel cation binding(GO:0016151) |
| 2.1 | 39.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.7 | 13.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 1.6 | 4.9 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 1.5 | 10.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 1.3 | 5.3 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 1.3 | 41.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 1.2 | 9.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.2 | 8.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.2 | 28.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 1.2 | 27.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 1.2 | 4.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.1 | 5.6 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 1.1 | 28.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 1.1 | 50.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.9 | 12.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.9 | 13.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.9 | 17.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.9 | 11.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.9 | 5.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.9 | 101.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.8 | 9.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.8 | 8.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.8 | 41.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.8 | 4.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.7 | 12.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.7 | 3.7 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.7 | 4.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.7 | 11.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.7 | 3.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.7 | 3.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.6 | 7.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.6 | 8.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.6 | 2.9 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.6 | 1.7 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.6 | 4.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.6 | 7.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.5 | 4.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.5 | 13.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.5 | 14.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.5 | 13.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.5 | 9.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.5 | 8.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.5 | 8.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.4 | 14.8 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.4 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.4 | 4.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.4 | 15.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 5.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.4 | 2.8 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.4 | 1.9 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.4 | 10.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 6.4 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.3 | 1.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 11.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 20.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.3 | 6.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 3.0 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) protein-hormone receptor activity(GO:0016500) |
| 0.2 | 1.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 2.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.2 | 13.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 26.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 5.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 2.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 3.0 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 0.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 28.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 1.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 19.1 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.2 | 39.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.2 | 41.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.2 | 5.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 19.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.2 | 0.5 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 6.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 30.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 25.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 1.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 27.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 35.1 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.1 | 3.8 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 7.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 2.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 1.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 2.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.7 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 3.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 4.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 4.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.7 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.1 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 2.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 5.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 3.4 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 0.7 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 45.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 1.4 | 27.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 1.3 | 9.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.9 | 17.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.9 | 72.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.9 | 40.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.8 | 9.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.8 | 6.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.7 | 31.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.7 | 29.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.6 | 8.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.5 | 4.9 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.5 | 15.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.5 | 7.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.5 | 9.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.5 | 13.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.5 | 22.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.5 | 19.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.4 | 3.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 20.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.4 | 7.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 7.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.3 | 4.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 5.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 15.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 8.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 12.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.2 | 4.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 8.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 43.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 8.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 6.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 41.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 25.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 13.5 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 6.7 | 261.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 4.3 | 38.7 | REACTOME DEFENSINS | Genes involved in Defensins |
| 2.2 | 43.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 1.8 | 64.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.7 | 22.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 1.2 | 50.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 1.1 | 10.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 1.0 | 41.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.9 | 21.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.8 | 17.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.8 | 9.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.7 | 8.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.7 | 11.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.6 | 15.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.4 | 8.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.4 | 9.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.4 | 5.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.4 | 4.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.4 | 47.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.4 | 29.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.4 | 10.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.4 | 2.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.4 | 8.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 3.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 10.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.3 | 64.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.3 | 4.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.3 | 4.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 9.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 5.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 4.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 1.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 3.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 10.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 6.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 8.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 3.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 5.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 4.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 3.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 2.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 1.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 0.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.7 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 9.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 2.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |