Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Tcf21_Msc
Z-value: 1.04
Transcription factors associated with Tcf21_Msc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
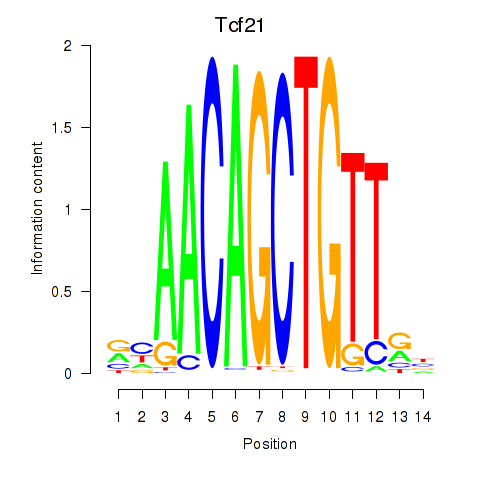
Tcf21
|
ENSRNOG00000016700 | transcription factor 21 |
|
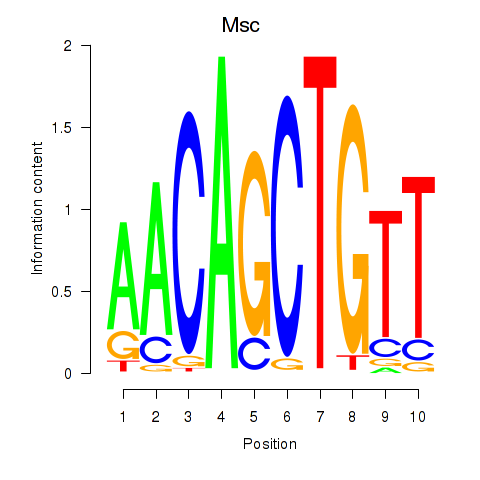
Msc
|
ENSRNOG00000007540 | musculin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msc | rn6_v1_chr5_+_3955227_3955227 | 0.37 | 1.5e-11 | Click! |
| Tcf21 | rn6_v1_chr1_+_23906717_23906717 | -0.10 | 6.2e-02 | Click! |
Activity profile of Tcf21_Msc motif
Sorted Z-values of Tcf21_Msc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_127248372 | 66.77 |
ENSRNOT00000085517
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr4_+_144382945 | 61.11 |
ENSRNOT00000007601
|
Cav3
|
caveolin 3 |
| chr1_+_80321585 | 52.40 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr15_-_33656089 | 51.42 |
ENSRNOT00000024186
|
Myh7
|
myosin heavy chain 7 |
| chr10_+_92628356 | 40.50 |
ENSRNOT00000072480
|
Myl4
|
myosin, light chain 4 |
| chr13_-_84331905 | 35.82 |
ENSRNOT00000004965
|
Dusp27
|
dual specificity phosphatase 27 (putative) |
| chr13_-_52197205 | 30.11 |
ENSRNOT00000009712
|
Shisa4
|
shisa family member 4 |
| chr11_+_36555416 | 29.22 |
ENSRNOT00000064981
|
Sh3bgr
|
SH3 domain binding glutamate-rich protein |
| chr2_-_45518502 | 27.72 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chrX_+_71342775 | 27.22 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr16_-_79671719 | 27.02 |
ENSRNOT00000015908
|
Myom2
|
myomesin 2 |
| chr11_-_39448503 | 25.31 |
ENSRNOT00000047347
|
LOC102553613
|
SH3 domain-binding glutamic acid-rich protein-like |
| chr19_+_14835822 | 24.93 |
ENSRNOT00000072804
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene |
| chr18_+_17043903 | 23.83 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr9_+_14529218 | 22.15 |
ENSRNOT00000016893
|
Apobec2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| chr6_-_26624092 | 21.89 |
ENSRNOT00000008113
|
Trim54
|
tripartite motif-containing 54 |
| chr1_+_201055644 | 21.86 |
ENSRNOT00000054937
ENSRNOT00000047161 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr13_+_80517536 | 21.14 |
ENSRNOT00000004386
|
Myoc
|
myocilin |
| chr9_-_43116521 | 20.81 |
ENSRNOT00000039437
|
Ankrd23
|
ankyrin repeat domain 23 |
| chr10_-_94557764 | 20.40 |
ENSRNOT00000016841
|
Scn4a
|
sodium voltage-gated channel alpha subunit 4 |
| chr6_-_60124274 | 19.23 |
ENSRNOT00000059823
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr8_+_53295222 | 18.75 |
ENSRNOT00000009724
ENSRNOT00000067420 |
Usp28
|
ubiquitin specific peptidase 28 |
| chr7_+_120153184 | 18.05 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr9_+_23503236 | 17.98 |
ENSRNOT00000017996
|
Crisp2
|
cysteine-rich secretory protein 2 |
| chr14_-_86047162 | 17.36 |
ENSRNOT00000018227
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr3_+_143129248 | 15.49 |
ENSRNOT00000006667
|
Cst8
|
cystatin 8 |
| chr13_+_52662996 | 15.40 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr1_-_254735548 | 15.30 |
ENSRNOT00000025258
|
Ankrd1
|
ankyrin repeat domain 1 |
| chr1_+_85112834 | 15.29 |
ENSRNOT00000026107
|
Dyrk1b
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr17_-_55346279 | 15.01 |
ENSRNOT00000025037
|
Svil
|
supervillin |
| chrX_-_77570825 | 15.01 |
ENSRNOT00000092505
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chrX_+_109996163 | 14.87 |
ENSRNOT00000093349
|
Nrk
|
Nik related kinase |
| chr1_-_143398093 | 14.66 |
ENSRNOT00000078916
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr14_-_92077088 | 14.28 |
ENSRNOT00000085175
|
Grb10
|
growth factor receptor bound protein 10 |
| chr16_-_8885797 | 14.22 |
ENSRNOT00000073370
|
RGD1564899
|
similar to chromosome 10 open reading frame 71 |
| chr13_+_85818427 | 14.22 |
ENSRNOT00000077227
ENSRNOT00000006117 |
Rxrg
|
retinoid X receptor gamma |
| chr1_-_199624783 | 14.01 |
ENSRNOT00000026908
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr15_+_34251606 | 13.83 |
ENSRNOT00000025725
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr4_-_60358562 | 13.73 |
ENSRNOT00000018001
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr13_+_52889737 | 13.61 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr16_+_6962722 | 13.57 |
ENSRNOT00000023330
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr16_-_49574314 | 13.42 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr3_+_11114551 | 13.27 |
ENSRNOT00000013507
|
Plpp7
|
phospholipid phosphatase 7 |
| chr12_+_42097626 | 13.07 |
ENSRNOT00000001893
|
Tbx5
|
T-box 5 |
| chr6_+_26602144 | 13.01 |
ENSRNOT00000008037
|
Ucn
|
urocortin |
| chr1_-_197858016 | 12.74 |
ENSRNOT00000074778
|
Atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr2_-_184289126 | 12.70 |
ENSRNOT00000081678
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr14_-_84106997 | 12.61 |
ENSRNOT00000065501
|
Osbp2
|
oxysterol binding protein 2 |
| chr12_+_30450316 | 12.36 |
ENSRNOT00000001222
|
Phkg1
|
phosphorylase kinase, gamma 1 |
| chr18_+_56180089 | 12.24 |
ENSRNOT00000030966
|
Arsi
|
arylsulfatase family, member I |
| chr1_-_107373807 | 12.17 |
ENSRNOT00000056024
|
Svip
|
small VCP interacting protein |
| chr7_-_80796670 | 12.11 |
ENSRNOT00000010539
|
Abra
|
actin-binding Rho activating protein |
| chr10_+_89251370 | 11.77 |
ENSRNOT00000076820
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr3_-_61494778 | 11.64 |
ENSRNOT00000068018
|
Lnpk
|
lunapark, ER junction formation factor |
| chr10_-_95934345 | 11.62 |
ENSRNOT00000004349
|
Cacng1
|
calcium voltage-gated channel auxiliary subunit gamma 1 |
| chr8_-_17525906 | 11.48 |
ENSRNOT00000007855
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr4_-_41212072 | 11.44 |
ENSRNOT00000085596
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr2_+_198683159 | 11.38 |
ENSRNOT00000028793
|
Txnip
|
thioredoxin interacting protein |
| chr17_+_15966234 | 11.36 |
ENSRNOT00000084304
|
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr16_-_40025401 | 11.30 |
ENSRNOT00000066639
|
Asb5
|
ankyrin repeat and SOCS box-containing 5 |
| chr10_-_8498422 | 11.24 |
ENSRNOT00000082332
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr18_+_14471213 | 11.14 |
ENSRNOT00000045224
|
Dtna
|
dystrobrevin, alpha |
| chr13_+_47602692 | 10.89 |
ENSRNOT00000038822
|
Fcmr
|
Fc fragment of IgM receptor |
| chr7_+_136037230 | 10.84 |
ENSRNOT00000071423
|
AABR07058745.1
|
|
| chrX_-_106066227 | 10.65 |
ENSRNOT00000033752
|
Nxf7
|
nuclear RNA export factor 7 |
| chr5_+_151692108 | 10.55 |
ENSRNOT00000086144
|
Fam46b
|
family with sequence similarity 46, member B |
| chr12_+_39553903 | 10.47 |
ENSRNOT00000001738
|
Atp2a2
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 |
| chr17_-_65955606 | 10.42 |
ENSRNOT00000067949
ENSRNOT00000023601 |
Ryr2
|
ryanodine receptor 2 |
| chr20_+_31102476 | 10.32 |
ENSRNOT00000078719
|
Lrrc20
|
leucine rich repeat containing 20 |
| chr5_+_157282669 | 10.26 |
ENSRNOT00000022827
|
Pla2g2a
|
phospholipase A2 group IIA |
| chr9_-_38196273 | 9.78 |
ENSRNOT00000044452
|
Dst
|
dystonin |
| chr5_-_168734296 | 9.78 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr1_+_90729274 | 9.46 |
ENSRNOT00000047207
|
RGD1560088
|
similar to NADH:ubiquinone oxidoreductase B15 subunit |
| chr10_+_54156649 | 9.37 |
ENSRNOT00000074718
|
Gas7
|
growth arrest specific 7 |
| chr13_+_50873605 | 9.33 |
ENSRNOT00000004382
|
Fmod
|
fibromodulin |
| chr15_-_34612432 | 9.20 |
ENSRNOT00000090206
|
Mcpt1l1
|
mast cell protease 1-like 1 |
| chr4_-_174180729 | 9.07 |
ENSRNOT00000011376
|
Plcz1
|
phospholipase C, zeta 1 |
| chr15_-_61772516 | 9.07 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chr7_+_95074236 | 8.84 |
ENSRNOT00000067649
|
Col14a1
|
collagen type XIV alpha 1 chain |
| chr6_+_55465782 | 8.77 |
ENSRNOT00000006682
|
Agr2
|
anterior gradient 2, protein disulphide isomerase family member |
| chr1_+_79989019 | 8.69 |
ENSRNOT00000020428
|
Dmpk
|
dystrophia myotonica-protein kinase |
| chr10_-_51778939 | 8.57 |
ENSRNOT00000078675
ENSRNOT00000057562 |
Myocd
|
myocardin |
| chr3_+_148386189 | 8.54 |
ENSRNOT00000011255
|
Mylk2
|
myosin light chain kinase 2 |
| chrX_+_109940350 | 8.22 |
ENSRNOT00000093543
ENSRNOT00000057049 |
Nrk
|
Nik related kinase |
| chr1_-_215846911 | 8.20 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr12_+_39554171 | 8.17 |
ENSRNOT00000024347
|
Atp2a2
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 |
| chr14_+_37113210 | 8.07 |
ENSRNOT00000089094
|
Sgcb
|
sarcoglycan, beta |
| chr20_-_33323367 | 8.05 |
ENSRNOT00000080444
|
Ros1
|
ROS proto-oncogene 1 , receptor tyrosine kinase |
| chr7_+_143092511 | 7.87 |
ENSRNOT00000084223
|
RGD1305207
|
similar to RIKEN cDNA 1700011A15 |
| chr13_-_51955735 | 7.76 |
ENSRNOT00000007597
|
Ptprv
|
protein tyrosine phosphatase, receptor type, V |
| chr4_+_158224000 | 7.74 |
ENSRNOT00000084240
ENSRNOT00000078495 |
Ano2
|
anoctamin 2 |
| chrX_+_106230013 | 7.61 |
ENSRNOT00000067828
|
AABR07040617.1
|
|
| chr4_+_14109864 | 7.61 |
ENSRNOT00000076349
|
RGD1565355
|
similar to fatty acid translocase/CD36 |
| chr10_-_89338739 | 7.55 |
ENSRNOT00000073923
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chrX_+_115721251 | 7.51 |
ENSRNOT00000060090
|
Trpc5os
|
TRPC5 opposite strand |
| chr20_+_5050327 | 7.42 |
ENSRNOT00000083353
|
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr9_-_115850634 | 7.37 |
ENSRNOT00000049720
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr10_-_107376645 | 7.26 |
ENSRNOT00000046213
|
Cep295nl
|
CEP295 N-terminal like |
| chrX_-_14783792 | 6.88 |
ENSRNOT00000087609
|
LOC691895
|
similar to ferritin, heavy polypeptide-like 17 |
| chr1_+_106405002 | 6.85 |
ENSRNOT00000074027
|
LOC103690168
|
probable isocitrate dehydrogenase [NAD] gamma 2, mitochondrial |
| chr10_-_97896525 | 6.85 |
ENSRNOT00000005172
|
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr5_-_160179978 | 6.73 |
ENSRNOT00000022820
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr10_-_51669297 | 6.68 |
ENSRNOT00000071595
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr2_-_219842986 | 6.67 |
ENSRNOT00000055735
|
Agl
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr20_+_28572242 | 6.63 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr20_-_33322966 | 6.57 |
ENSRNOT00000000459
|
Ros1
|
ROS proto-oncogene 1 , receptor tyrosine kinase |
| chr6_-_88232252 | 6.57 |
ENSRNOT00000047597
|
Rpl10l
|
ribosomal protein L10-like |
| chr11_-_36533073 | 6.57 |
ENSRNOT00000033486
|
Lca5l
|
LCA5L, lebercilin like |
| chrX_+_106002416 | 6.54 |
ENSRNOT00000052196
|
LOC100910698
|
leucine-rich repeat-containing protein PRAME-like |
| chr7_-_11777503 | 6.48 |
ENSRNOT00000026220
|
Amh
|
anti-Mullerian hormone |
| chr1_+_162890475 | 6.46 |
ENSRNOT00000034886
|
Gdpd4
|
glycerophosphodiester phosphodiesterase domain containing 4 |
| chr12_+_19561347 | 6.42 |
ENSRNOT00000060025
|
RGD1562319
|
similar to family with sequence similarity 55, member C |
| chr3_+_172195844 | 6.39 |
ENSRNOT00000034915
|
Npepl1
|
aminopeptidase-like 1 |
| chr5_-_64946580 | 6.38 |
ENSRNOT00000007290
|
Ppp3r2
|
protein phosphatase 3, regulatory subunit B, beta |
| chr11_-_87434482 | 6.37 |
ENSRNOT00000074346
ENSRNOT00000002557 |
Lrrc74b
|
leucine rich repeat containing 74B |
| chr18_-_56870801 | 6.21 |
ENSRNOT00000087188
ENSRNOT00000032189 |
Arhgef37
|
Rho guanine nucleotide exchange factor 37 |
| chr3_-_5617225 | 6.15 |
ENSRNOT00000008327
|
Tmem8c
|
transmembrane protein 8C |
| chrX_-_39956841 | 6.13 |
ENSRNOT00000050708
|
Klhl34
|
kelch-like family member 34 |
| chr8_-_49038169 | 6.07 |
ENSRNOT00000047303
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr16_-_12124265 | 6.02 |
ENSRNOT00000080192
|
AABR07024682.1
|
|
| chr3_-_141305471 | 6.00 |
ENSRNOT00000016915
|
Nkx2-4
|
NK2 homeobox 4 |
| chr10_+_16542882 | 5.89 |
ENSRNOT00000028146
|
Stc2
|
stanniocalcin 2 |
| chr7_+_144565392 | 5.82 |
ENSRNOT00000021589
|
Hoxc11
|
homeobox C11 |
| chr4_-_157798868 | 5.82 |
ENSRNOT00000044425
|
Tuba3b
|
tubulin, alpha 3B |
| chr9_+_42871950 | 5.82 |
ENSRNOT00000089673
|
Arid5a
|
AT-rich interaction domain 5A |
| chr3_-_148057523 | 5.81 |
ENSRNOT00000055408
|
Defb24
|
defensin beta 24 |
| chr5_-_17399801 | 5.76 |
ENSRNOT00000011900
|
RGD1563405
|
similar to protein tyrosine phosphatase 4a2 |
| chr13_+_56262190 | 5.74 |
ENSRNOT00000032908
|
AABR07021086.1
|
|
| chr12_-_18227991 | 5.68 |
ENSRNOT00000072986
|
AABR07035501.1
|
|
| chr10_-_43891885 | 5.56 |
ENSRNOT00000084984
|
LOC691286
|
similar to RIKEN cDNA 4930504O13 |
| chr16_+_13050305 | 5.54 |
ENSRNOT00000077388
|
AABR07024735.1
|
|
| chr4_+_166833320 | 5.47 |
ENSRNOT00000031438
|
Tas2r120
|
taste receptor, type 2, member 120 |
| chr18_-_55891710 | 5.42 |
ENSRNOT00000064686
|
Synpo
|
synaptopodin |
| chr9_+_82571269 | 5.34 |
ENSRNOT00000026941
|
Speg
|
SPEG complex locus |
| chr8_+_64489942 | 5.31 |
ENSRNOT00000083666
|
Pkm
|
pyruvate kinase, muscle |
| chrX_+_62366453 | 5.24 |
ENSRNOT00000089828
|
Arx
|
aristaless related homeobox |
| chrX_-_66602458 | 5.13 |
ENSRNOT00000076510
ENSRNOT00000017429 |
Eda2r
|
ectodysplasin A2 receptor |
| chr3_+_138174054 | 5.12 |
ENSRNOT00000007946
|
Banf2
|
barrier to autointegration factor 2 |
| chr5_+_147692391 | 5.11 |
ENSRNOT00000064855
|
Fam229a
|
family with sequence similarity 229, member A |
| chr1_+_85112247 | 5.09 |
ENSRNOT00000087157
|
Dyrk1b
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr5_+_90419020 | 5.05 |
ENSRNOT00000058893
|
Rasef
|
RAS and EF hand domain containing |
| chr20_-_13647581 | 5.02 |
ENSRNOT00000083190
|
RGD1563815
|
similar to sodium-glucose cotransporter-like 1 |
| chr2_-_172361779 | 5.02 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr1_-_227506822 | 5.01 |
ENSRNOT00000091506
|
Ms4a7
|
membrane spanning 4-domains A7 |
| chr9_-_89193821 | 5.01 |
ENSRNOT00000090881
|
Sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr1_+_264827739 | 4.98 |
ENSRNOT00000021553
|
Kazald1
|
Kazal-type serine peptidase inhibitor domain 1 |
| chr7_-_117712213 | 4.94 |
ENSRNOT00000092930
|
Cyhr1
|
cysteine and histidine rich 1 |
| chr12_-_24365324 | 4.92 |
ENSRNOT00000032250
|
Trim50
|
tripartite motif-containing 50 |
| chr15_+_52767442 | 4.90 |
ENSRNOT00000014441
|
LOC108348161
|
phytanoyl-CoA hydroxylase-interacting protein |
| chr2_+_189955836 | 4.73 |
ENSRNOT00000078351
|
S100a3
|
S100 calcium binding protein A3 |
| chr14_+_91588940 | 4.72 |
ENSRNOT00000057067
|
RGD1309870
|
hypothetical LOC289778 |
| chrX_-_38196060 | 4.71 |
ENSRNOT00000006741
ENSRNOT00000006438 |
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr14_-_89179507 | 4.67 |
ENSRNOT00000006498
|
Pkd1l1
|
polycystin 1 like 1, transient receptor potential channel interacting |
| chr7_+_40316639 | 4.65 |
ENSRNOT00000080150
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr15_+_4240203 | 4.62 |
ENSRNOT00000010178
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr16_+_12174370 | 4.58 |
ENSRNOT00000072045
|
LOC102553538
|
ral guanine nucleotide dissociation stimulator-like |
| chr1_+_53531076 | 4.54 |
ENSRNOT00000018015
|
Tcp10b
|
t-complex protein 10b |
| chr1_-_227359809 | 4.48 |
ENSRNOT00000088080
|
1700025F22Rik
|
RIKEN cDNA 1700025F22 gene |
| chr4_+_1841955 | 4.48 |
ENSRNOT00000019271
|
Olr1096
|
olfactory receptor 1096 |
| chr3_+_149606940 | 4.41 |
ENSRNOT00000018512
|
Bpifa3
|
BPI fold containing family A, member 3 |
| chrX_-_111942749 | 4.38 |
ENSRNOT00000087583
|
AABR07040840.1
|
|
| chr2_-_205212681 | 4.37 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr13_-_91776397 | 4.36 |
ENSRNOT00000073147
|
Mptx1
|
mucosal pentraxin 1 |
| chr2_+_243840134 | 4.36 |
ENSRNOT00000093355
|
Eif4e
|
eukaryotic translation initiation factor 4E |
| chrX_+_25737292 | 4.35 |
ENSRNOT00000004893
|
RGD1563947
|
similar to RIKEN cDNA 4933400A11 |
| chr2_+_92559929 | 4.35 |
ENSRNOT00000033404
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr9_-_114282799 | 4.33 |
ENSRNOT00000090539
|
LOC102555328
|
uncharacterized LOC102555328 |
| chr7_-_60743328 | 4.26 |
ENSRNOT00000066767
|
Mdm2
|
MDM2 proto-oncogene |
| chr10_-_98469799 | 4.24 |
ENSRNOT00000087502
ENSRNOT00000088646 |
Abca9
|
ATP binding cassette subfamily A member 9 |
| chr2_+_210381829 | 4.22 |
ENSRNOT00000024611
|
Alx3
|
ALX homeobox 3 |
| chr2_-_123851854 | 4.22 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr8_+_116343096 | 4.22 |
ENSRNOT00000022092
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr8_-_43336304 | 4.20 |
ENSRNOT00000036054
|
RGD1311744
|
similar to RIKEN cDNA 5830475I06 |
| chr19_+_25123724 | 4.20 |
ENSRNOT00000007407
|
LOC686013
|
hypothetical protein LOC686013 |
| chr16_-_12626949 | 4.20 |
ENSRNOT00000077469
|
AABR07024722.1
|
|
| chr7_-_130368819 | 4.11 |
ENSRNOT00000064328
|
Syce3
|
synaptonemal complex central element protein 3 |
| chr17_+_60287203 | 4.08 |
ENSRNOT00000025585
|
Armc4
|
armadillo repeat containing 4 |
| chrX_+_63520991 | 4.03 |
ENSRNOT00000071590
|
Apoo
|
apolipoprotein O |
| chr17_-_72046381 | 4.00 |
ENSRNOT00000073919
|
AABR07028488.1
|
|
| chr14_-_38575785 | 3.99 |
ENSRNOT00000003146
|
Atp10d
|
ATPase phospholipid transporting 10D (putative) |
| chr13_+_83681322 | 3.93 |
ENSRNOT00000004206
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr19_-_62158327 | 3.93 |
ENSRNOT00000075053
|
Ccdc7
|
coiled-coil domain containing 7 |
| chr8_-_17833773 | 3.91 |
ENSRNOT00000049758
|
Olr1117
|
olfactory receptor 1117 |
| chr9_+_69790831 | 3.89 |
ENSRNOT00000045156
|
LOC100360781
|
ribosomal protein L37-like |
| chr2_+_187740531 | 3.88 |
ENSRNOT00000092653
|
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr1_+_141218095 | 3.87 |
ENSRNOT00000051411
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr1_+_220446425 | 3.86 |
ENSRNOT00000027339
|
LOC100911932
|
endosialin-like |
| chr1_+_220362064 | 3.86 |
ENSRNOT00000074361
|
LOC108348067
|
endosialin |
| chr3_+_150910398 | 3.82 |
ENSRNOT00000055310
|
Tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr9_-_80166807 | 3.82 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr9_+_51009116 | 3.82 |
ENSRNOT00000039313
|
Mettl21cl1
|
methyltransferase like 21C-like 1 |
| chr9_-_120101098 | 3.81 |
ENSRNOT00000002917
|
Fam114a1l1
|
family with sequence similarity 114, member A1-like 1 |
| chr19_+_755460 | 3.81 |
ENSRNOT00000076560
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chr1_-_221281180 | 3.81 |
ENSRNOT00000028379
|
Cdc42ep2
|
CDC42 effector protein 2 |
| chr7_+_123102493 | 3.73 |
ENSRNOT00000038612
|
Aco2
|
aconitase 2 |
| chr3_+_148047638 | 3.72 |
ENSRNOT00000031613
|
Defb21
|
defensin beta 21 |
| chr2_-_52282548 | 3.72 |
ENSRNOT00000033627
|
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr13_+_50434702 | 3.72 |
ENSRNOT00000032244
|
Sox13
|
SRY box 13 |
| chr14_+_43512901 | 3.72 |
ENSRNOT00000050664
|
Apbb2
|
amyloid beta precursor protein binding family B member 2 |
| chrX_+_122938009 | 3.70 |
ENSRNOT00000089266
ENSRNOT00000017550 |
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf21_Msc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.2 | 72.7 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 15.3 | 61.1 | GO:2001288 | detection of muscle stretch(GO:0035995) positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 9.3 | 18.6 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) |
| 5.8 | 40.6 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 5.5 | 22.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 5.3 | 21.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) ERBB3 signaling pathway(GO:0038129) |
| 5.1 | 15.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 4.3 | 13.0 | GO:0035483 | gastric emptying(GO:0035483) |
| 4.2 | 12.7 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 4.1 | 12.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 3.8 | 23.1 | GO:0060723 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 3.6 | 18.0 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 3.5 | 10.4 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 3.4 | 13.6 | GO:1902730 | positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 3.0 | 12.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 2.9 | 8.6 | GO:2000721 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 2.6 | 20.4 | GO:0015871 | choline transport(GO:0015871) |
| 2.5 | 17.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 2.4 | 14.6 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 2.3 | 20.8 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 2.2 | 13.1 | GO:0003166 | bundle of His development(GO:0003166) positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 2.2 | 6.5 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 2.1 | 4.3 | GO:1904404 | response to formaldehyde(GO:1904404) |
| 2.0 | 6.1 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 2.0 | 14.3 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 2.0 | 13.8 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.7 | 8.7 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 1.7 | 6.9 | GO:0031179 | peptide modification(GO:0031179) |
| 1.7 | 23.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 1.7 | 11.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.5 | 8.8 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 1.5 | 21.9 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 1.4 | 5.6 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 1.4 | 40.7 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 1.3 | 3.9 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 1.2 | 8.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.2 | 14.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.1 | 14.5 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 1.1 | 17.8 | GO:0042407 | cristae formation(GO:0042407) |
| 1.1 | 3.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 1.1 | 4.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 1.0 | 8.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 1.0 | 8.9 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 1.0 | 10.9 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 1.0 | 2.9 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.0 | 3.8 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.9 | 76.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.9 | 2.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.9 | 2.8 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.9 | 7.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.9 | 2.8 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.9 | 2.8 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.9 | 9.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.9 | 48.9 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.9 | 11.5 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.9 | 5.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.8 | 2.5 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.8 | 3.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.8 | 1.5 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.8 | 13.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.7 | 4.2 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.7 | 11.8 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.7 | 23.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.7 | 2.6 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.6 | 6.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.6 | 13.3 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.6 | 3.7 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.6 | 8.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.6 | 11.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.6 | 3.5 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.6 | 8.7 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.6 | 14.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.6 | 2.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.5 | 11.4 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.5 | 2.7 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.5 | 5.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.5 | 5.8 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.5 | 6.9 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.5 | 1.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.5 | 6.8 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.5 | 2.3 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.4 | 19.3 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.4 | 11.6 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.4 | 12.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.4 | 9.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.4 | 2.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 8.4 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.4 | 12.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.4 | 1.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.4 | 3.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.4 | 5.9 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.4 | 18.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.4 | 1.2 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.4 | 19.5 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.3 | 3.8 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.3 | 1.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.3 | 2.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 4.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.3 | 2.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 | 0.6 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.3 | 5.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 7.0 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.3 | 0.3 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.3 | 0.9 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.3 | 2.0 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.3 | 3.7 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.2 | 0.5 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 6.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 1.7 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 6.5 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.2 | 2.6 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.2 | 1.9 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.2 | 3.7 | GO:0048535 | lymph node development(GO:0048535) |
| 0.2 | 0.9 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.2 | 1.2 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.2 | 3.0 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.2 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 0.2 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.2 | 0.7 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 4.2 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.2 | 0.9 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 2.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 2.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.2 | 2.3 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.2 | 17.9 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.2 | 2.3 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.2 | 5.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 1.0 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.2 | 0.5 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.2 | 1.5 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 4.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.2 | 0.9 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.2 | 0.9 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 1.2 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.1 | 0.7 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 2.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 1.2 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.6 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.1 | 0.9 | GO:0099525 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 14.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.7 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 0.2 | GO:1904528 | positive regulation of microtubule binding(GO:1904528) |
| 0.1 | 27.8 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.1 | 0.5 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 1.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 6.1 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.1 | 0.3 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.6 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.1 | 0.3 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.9 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 1.0 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.9 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.9 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.9 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.6 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.1 | 0.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.4 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 13.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 2.6 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 0.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.7 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 1.2 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 0.6 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.3 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 2.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.7 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 | 0.8 | GO:1900040 | regulation of interleukin-2 secretion(GO:1900040) |
| 0.1 | 0.7 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 0.7 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 2.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.8 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 2.2 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 1.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 1.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 3.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 0.6 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 1.0 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 2.1 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 1.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.0 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 3.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.3 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 1.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 3.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.8 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0060383 | positive regulation of DNA strand elongation(GO:0060383) |
| 0.0 | 3.0 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 9.3 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.4 | GO:0050718 | positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 0.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.8 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.5 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.4 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 4.8 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.4 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.6 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.4 | GO:0098840 | intraciliary transport(GO:0042073) protein transport along microtubule(GO:0098840) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 66.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 7.0 | 21.1 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 6.5 | 78.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 6.2 | 18.6 | GO:0090534 | longitudinal sarcoplasmic reticulum(GO:0014801) calcium ion-transporting ATPase complex(GO:0090534) |
| 3.8 | 15.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 3.3 | 62.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 2.5 | 12.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 2.5 | 17.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 2.4 | 12.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 2.0 | 9.8 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 1.8 | 5.4 | GO:0097444 | spine apparatus(GO:0097444) |
| 1.5 | 12.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 1.3 | 62.8 | GO:0031672 | A band(GO:0031672) |
| 1.1 | 3.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.1 | 23.8 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 1.0 | 20.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.9 | 16.9 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.9 | 6.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.8 | 15.0 | GO:0043034 | costamere(GO:0043034) |
| 0.8 | 98.7 | GO:0031674 | I band(GO:0031674) |
| 0.7 | 13.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.6 | 12.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.6 | 2.8 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.6 | 8.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.5 | 6.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.5 | 4.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.5 | 10.5 | GO:0030017 | sarcomere(GO:0030017) |
| 0.5 | 8.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 2.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.4 | 1.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.4 | 7.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 1.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.4 | 2.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 16.3 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.3 | 1.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 1.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 4.1 | GO:0000801 | central element(GO:0000801) |
| 0.3 | 2.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 1.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 7.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 0.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 4.0 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 2.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 0.6 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.2 | 2.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 4.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 7.6 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 3.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 10.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.6 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 5.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.7 | GO:0097169 | NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 2.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 3.9 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 4.1 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 2.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.9 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 2.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 0.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 6.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 34.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 1.8 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 11.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 0.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 2.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 6.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.4 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.2 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 0.1 | 2.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 2.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 6.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 6.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 5.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 15.4 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 1.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 1.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 5.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 8.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 11.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 4.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.7 | GO:0044438 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 61.1 | GO:0071253 | connexin binding(GO:0071253) |
| 7.5 | 52.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 4.9 | 29.7 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 4.3 | 17.4 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 3.8 | 15.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 3.7 | 22.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 3.4 | 48.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 3.3 | 13.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 3.0 | 18.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 2.8 | 16.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 2.6 | 10.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 2.6 | 36.1 | GO:0031432 | titin binding(GO:0031432) |
| 2.5 | 12.7 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 2.5 | 7.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 2.1 | 12.7 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 2.1 | 27.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.7 | 6.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.7 | 6.9 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 1.5 | 12.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 1.3 | 5.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.2 | 3.7 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 1.2 | 13.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 1.2 | 18.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 1.2 | 6.9 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 1.1 | 10.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 1.1 | 11.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.9 | 6.4 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.9 | 12.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.8 | 43.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.7 | 21.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.7 | 13.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.7 | 4.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.7 | 2.8 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.7 | 20.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.7 | 2.7 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.7 | 4.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.6 | 3.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.6 | 2.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.6 | 16.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.5 | 7.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.5 | 8.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.5 | 5.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.5 | 1.9 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.5 | 4.2 | GO:0043208 | opioid receptor binding(GO:0031628) glycosphingolipid binding(GO:0043208) |
| 0.5 | 12.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.5 | 10.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.5 | 8.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.4 | 13.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 1.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.4 | 9.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.4 | 4.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.4 | 11.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.4 | 5.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.4 | 7.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 1.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 1.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.3 | 13.0 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.3 | 69.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.3 | 6.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.3 | 8.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 5.8 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 8.1 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.3 | 1.0 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 2.2 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.2 | 20.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 6.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 0.9 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.2 | 17.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 0.9 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 0.6 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.2 | 2.6 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.2 | 1.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.2 | 1.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 0.7 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.2 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 0.7 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 22.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 10.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 2.9 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.2 | 23.0 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.2 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 0.9 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.2 | 0.9 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.2 | 6.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 12.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.7 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 2.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.6 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 0.1 | 2.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.7 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 3.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 5.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 7.0 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 1.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 4.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 4.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 11.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 1.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 0.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 5.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.7 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 5.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.4 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.2 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.1 | 0.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 5.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 4.1 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 3.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 0.9 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 0.7 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.1 | 5.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 6.1 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 0.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 11.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 10.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 1.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 7.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 3.2 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 1.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 5.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.6 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 61.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.9 | 10.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.7 | 2.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.5 | 14.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.5 | 15.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.5 | 6.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.4 | 11.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.4 | 12.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.4 | 31.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.4 | 21.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.3 | 8.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 8.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 5.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 2.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 8.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 6.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 2.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.2 | 4.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 3.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 4.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 8.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 33.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 1.4 | 55.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.2 | 12.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 1.0 | 12.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.0 | 14.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 1.0 | 19.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.7 | 9.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.6 | 11.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.5 | 16.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 9.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 6.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.4 | 17.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 1.3 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.4 | 12.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.4 | 7.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.4 | 4.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.4 | 90.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.3 | 14.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 2.1 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.3 | 8.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.3 | 4.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 2.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 4.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 29.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 1.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 7.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 1.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 11.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 15.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.2 | 7.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 3.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 6.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.8 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 0.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 5.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.4 | REACTOME ORC1 REMOVAL FROM CHROMATIN | Genes involved in Orc1 removal from chromatin |
| 0.1 | 1.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.3 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.0 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 4.5 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.6 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.7 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.9 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |