Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
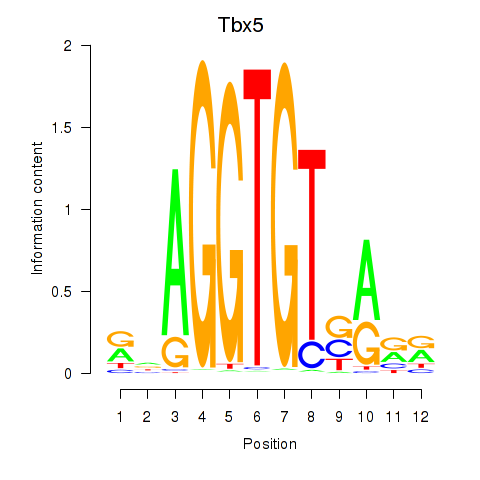
Results for Tbx5
Z-value: 0.97
Transcription factors associated with Tbx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx5
|
ENSRNOG00000001399 | T-box 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx5 | rn6_v1_chr12_+_42097626_42097626 | 0.39 | 2.9e-13 | Click! |
Activity profile of Tbx5 motif
Sorted Z-values of Tbx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_53609214 | 85.37 |
ENSRNOT00000077443
|
Slc27a6
|
solute carrier family 27 member 6 |
| chr6_-_2311781 | 29.87 |
ENSRNOT00000084171
|
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr8_+_62779875 | 27.24 |
ENSRNOT00000010831
|
Cyp11a1
|
cytochrome P450, family 11, subfamily a, polypeptide 1 |
| chr2_+_230901126 | 25.54 |
ENSRNOT00000016026
ENSRNOT00000015564 ENSRNOT00000068198 |
Camk2d
|
calcium/calmodulin-dependent protein kinase II delta |
| chr10_-_91661558 | 24.60 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr5_+_164808323 | 22.48 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr8_-_107952530 | 21.00 |
ENSRNOT00000052043
|
Cldn18
|
claudin 18 |
| chr10_+_107455845 | 20.51 |
ENSRNOT00000004373
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr1_+_156552328 | 19.38 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chrX_+_33443186 | 18.75 |
ENSRNOT00000005622
|
S100g
|
S100 calcium binding protein G |
| chr1_-_236729412 | 17.61 |
ENSRNOT00000054794
|
Prune2
|
prune homolog 2 |
| chr4_+_120133713 | 16.45 |
ENSRNOT00000017240
|
Gata2
|
GATA binding protein 2 |
| chr7_-_116255167 | 14.95 |
ENSRNOT00000038109
ENSRNOT00000041774 |
Cyp11b2
|
cytochrome P450, family 11, subfamily b, polypeptide 2 |
| chr2_-_189856090 | 14.64 |
ENSRNOT00000020307
|
Npr1
|
natriuretic peptide receptor 1 |
| chr10_-_56511583 | 13.72 |
ENSRNOT00000021402
|
LOC497940
|
similar to RIKEN cDNA 2810408A11 |
| chr20_+_44436403 | 13.41 |
ENSRNOT00000000733
ENSRNOT00000076859 |
Fyn
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr1_-_98521551 | 13.32 |
ENSRNOT00000081922
|
Siglec10
|
sialic acid binding Ig-like lectin 10 |
| chr5_+_157222636 | 13.29 |
ENSRNOT00000022579
|
Pla2g2d
|
phospholipase A2, group IID |
| chr4_-_115015965 | 12.99 |
ENSRNOT00000014603
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr2_+_93792601 | 12.52 |
ENSRNOT00000014701
ENSRNOT00000077311 |
Fabp4
|
fatty acid binding protein 4 |
| chr5_-_57896475 | 12.48 |
ENSRNOT00000017903
|
RGD1561916
|
similar to testes development-related NYD-SP22 isoform 1 |
| chr11_+_33863500 | 12.26 |
ENSRNOT00000072384
|
LOC102556347
|
carbonyl reductase [NADPH] 1-like |
| chr10_+_54156649 | 12.24 |
ENSRNOT00000074718
|
Gas7
|
growth arrest specific 7 |
| chr4_-_158010166 | 12.24 |
ENSRNOT00000026633
|
Cd9
|
CD9 molecule |
| chr11_+_33845463 | 12.23 |
ENSRNOT00000041838
|
Cbr1
|
carbonyl reductase 1 |
| chr1_+_100501676 | 12.22 |
ENSRNOT00000043724
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr11_+_33812989 | 12.00 |
ENSRNOT00000042283
ENSRNOT00000075985 |
Cbr1
|
carbonyl reductase 1 |
| chr1_+_143675985 | 11.92 |
ENSRNOT00000078500
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr1_+_213595240 | 11.89 |
ENSRNOT00000017137
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chrX_-_123601100 | 11.88 |
ENSRNOT00000092546
ENSRNOT00000092301 |
Sept6
|
septin 6 |
| chr6_-_138744480 | 11.86 |
ENSRNOT00000089387
|
AABR07065651.5
|
|
| chr1_-_98521706 | 11.74 |
ENSRNOT00000015941
|
Siglec10
|
sialic acid binding Ig-like lectin 10 |
| chr16_-_8564379 | 11.63 |
ENSRNOT00000060975
|
LOC680885
|
hypothetical protein LOC680885 |
| chr16_+_26906716 | 11.56 |
ENSRNOT00000064297
|
Cpe
|
carboxypeptidase E |
| chr7_-_12635943 | 11.09 |
ENSRNOT00000015029
|
Cfd
|
complement factor D |
| chr10_+_69434941 | 10.71 |
ENSRNOT00000009756
|
Ccl11
|
C-C motif chemokine ligand 11 |
| chr1_+_203160323 | 10.67 |
ENSRNOT00000027919
|
AABR07005844.1
|
|
| chr15_+_32894938 | 10.53 |
ENSRNOT00000012837
|
Abhd4
|
abhydrolase domain containing 4 |
| chr1_-_178195948 | 10.44 |
ENSRNOT00000081767
|
Btbd10
|
BTB domain containing 10 |
| chr3_-_164388492 | 10.20 |
ENSRNOT00000090493
|
Tmem189
|
transmembrane protein 189 |
| chr9_-_80167033 | 10.19 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr10_-_82887497 | 10.12 |
ENSRNOT00000005644
|
Itga3
|
integrin subunit alpha 3 |
| chr9_-_80166807 | 9.82 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr11_+_33812662 | 9.67 |
ENSRNOT00000085961
|
LOC102556347
|
carbonyl reductase [NADPH] 1-like |
| chr2_+_174013288 | 9.64 |
ENSRNOT00000013904
|
Serpini1
|
serpin family I member 1 |
| chr8_-_116361343 | 9.08 |
ENSRNOT00000066296
|
Sema3b
|
semaphorin 3B |
| chr1_-_53087474 | 9.06 |
ENSRNOT00000017302
|
Ccr6
|
C-C motif chemokine receptor 6 |
| chr6_-_137733026 | 9.03 |
ENSRNOT00000019213
|
Jag2
|
jagged 2 |
| chr5_+_151436464 | 8.80 |
ENSRNOT00000012463
|
Map3k6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr10_-_70735742 | 8.56 |
ENSRNOT00000077035
|
Heatr9
|
HEAT repeat containing 9 |
| chr1_-_271256379 | 8.44 |
ENSRNOT00000081605
|
LOC100912218
|
inositol 1,4,5-trisphosphate receptor-interacting protein-like |
| chr13_+_52662996 | 8.44 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr4_+_147275334 | 8.24 |
ENSRNOT00000051858
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
| chr13_+_110920830 | 8.24 |
ENSRNOT00000077014
ENSRNOT00000076362 |
Kcnh1
|
potassium voltage-gated channel subfamily H member 1 |
| chr19_+_12097632 | 8.15 |
ENSRNOT00000049452
|
Ces5a
|
carboxylesterase 5A |
| chr8_-_104995725 | 8.13 |
ENSRNOT00000037120
|
Slc25a36
|
solute carrier family 25 member 36 |
| chr3_-_11417546 | 8.08 |
ENSRNOT00000018776
|
Lcn2
|
lipocalin 2 |
| chr8_-_82533689 | 8.01 |
ENSRNOT00000014124
|
Tmod2
|
tropomodulin 2 |
| chr20_+_13498926 | 8.00 |
ENSRNOT00000070992
ENSRNOT00000045375 |
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chrX_-_123600890 | 7.98 |
ENSRNOT00000067942
|
Sept6
|
septin 6 |
| chr8_+_39997875 | 7.86 |
ENSRNOT00000050609
|
Esam
|
endothelial cell adhesion molecule |
| chr1_+_189870622 | 7.77 |
ENSRNOT00000075003
|
Tmem159
|
transmembrane protein 159 |
| chr20_+_3351303 | 7.65 |
ENSRNOT00000080419
ENSRNOT00000001065 ENSRNOT00000086503 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr1_+_100447998 | 7.63 |
ENSRNOT00000026259
|
Lrrc4b
|
leucine rich repeat containing 4B |
| chr7_-_62972084 | 7.56 |
ENSRNOT00000064251
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chr10_-_86688730 | 7.43 |
ENSRNOT00000055333
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr4_-_129515435 | 7.23 |
ENSRNOT00000039353
|
Eogt
|
EGF domain specific O-linked N-acetylglucosamine transferase |
| chr6_-_136145837 | 7.11 |
ENSRNOT00000015122
|
Ckb
|
creatine kinase B |
| chr9_+_100830250 | 7.02 |
ENSRNOT00000024526
|
Bok
|
BOK, BCL2 family apoptosis regulator |
| chr9_+_82596355 | 6.97 |
ENSRNOT00000065076
|
Speg
|
SPEG complex locus |
| chr4_-_119568736 | 6.95 |
ENSRNOT00000041234
|
Aplf
|
aprataxin and PNKP like factor |
| chr13_+_52147555 | 6.92 |
ENSRNOT00000084766
|
Lmod1
|
leiomodin 1 |
| chr5_-_58198782 | 6.86 |
ENSRNOT00000023951
|
Ccl21
|
C-C motif chemokine ligand 21 |
| chr15_+_62406873 | 6.76 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr11_+_33909439 | 6.71 |
ENSRNOT00000002310
|
Cbr3
|
carbonyl reductase 3 |
| chr16_-_62373253 | 6.69 |
ENSRNOT00000034325
|
Tex15
|
testis expressed 15, meiosis and synapsis associated |
| chr14_+_86514214 | 6.60 |
ENSRNOT00000052002
|
LOC103693776
|
zinc finger MIZ domain-containing protein 2 |
| chr2_-_3124543 | 6.60 |
ENSRNOT00000036547
|
Fam81b
|
family with sequence similarity 81, member B |
| chr1_-_80783898 | 6.58 |
ENSRNOT00000045306
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr15_+_87886783 | 6.56 |
ENSRNOT00000065710
|
Slain1
|
SLAIN motif family, member 1 |
| chr10_-_82887301 | 6.55 |
ENSRNOT00000035894
|
Itga3
|
integrin subunit alpha 3 |
| chr2_+_114386019 | 6.49 |
ENSRNOT00000082148
|
AABR07009834.1
|
|
| chr6_-_140572023 | 6.38 |
ENSRNOT00000072338
|
AABR07065772.1
|
|
| chr1_-_142500367 | 6.28 |
ENSRNOT00000015948
ENSRNOT00000089591 |
Crtc3
|
CREB regulated transcription coactivator 3 |
| chr20_-_3401273 | 6.23 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr8_+_116336441 | 6.17 |
ENSRNOT00000090123
ENSRNOT00000021590 |
Nat6
Hyal3
|
N-acetyltransferase 6 hyaluronoglucosaminidase 3 |
| chr3_+_80556668 | 6.11 |
ENSRNOT00000079118
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr1_+_235166718 | 6.09 |
ENSRNOT00000020201
|
Gna14
|
G protein subunit alpha 14 |
| chr2_-_186245163 | 6.09 |
ENSRNOT00000089339
|
Dclk2
|
doublecortin-like kinase 2 |
| chr7_+_120580743 | 6.05 |
ENSRNOT00000017181
|
Maff
|
MAF bZIP transcription factor F |
| chr5_-_6186329 | 6.01 |
ENSRNOT00000012610
|
Sulf1
|
sulfatase 1 |
| chr10_-_13107771 | 6.01 |
ENSRNOT00000005879
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr4_+_85551502 | 6.00 |
ENSRNOT00000087191
ENSRNOT00000015692 |
Aqp1
|
aquaporin 1 |
| chr4_+_147037179 | 5.99 |
ENSRNOT00000011292
|
Syn2
|
synapsin II |
| chr1_-_190914610 | 5.99 |
ENSRNOT00000023189
|
Cdr2
|
cerebellar degeneration-related protein 2 |
| chr17_-_86322144 | 5.98 |
ENSRNOT00000034474
|
LOC681241
|
hypothetical protein LOC681241 |
| chr1_+_141550633 | 5.78 |
ENSRNOT00000020047
|
Mesp2
|
mesoderm posterior bHLH transcription factor 2 |
| chr16_-_18645941 | 5.76 |
ENSRNOT00000079241
|
Dydc2
|
DPY30 domain containing 2 |
| chr2_-_59084059 | 5.75 |
ENSRNOT00000086323
ENSRNOT00000088701 |
Spef2
|
sperm flagellar 2 |
| chr15_+_57221292 | 5.63 |
ENSRNOT00000014502
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr6_-_139102378 | 5.61 |
ENSRNOT00000086423
|
AABR07065656.5
|
|
| chr18_-_49937522 | 5.60 |
ENSRNOT00000033254
|
Zfp608
|
zinc finger protein 608 |
| chr3_+_93920447 | 5.57 |
ENSRNOT00000012625
|
Lmo2
|
LIM domain only 2 |
| chr7_+_129595192 | 5.51 |
ENSRNOT00000071151
|
Zdhhc25
|
zinc finger, DHHC-type containing 25 |
| chr5_-_147691997 | 5.39 |
ENSRNOT00000011887
|
Tssk3
|
testis-specific serine kinase 3 |
| chr6_-_138550576 | 5.33 |
ENSRNOT00000075284
|
AABR07065645.1
|
|
| chr1_+_256786124 | 5.32 |
ENSRNOT00000034563
|
Ffar4
|
free fatty acid receptor 4 |
| chr4_+_142780987 | 5.31 |
ENSRNOT00000056570
|
Grm7
|
glutamate metabotropic receptor 7 |
| chr9_+_77320726 | 5.30 |
ENSRNOT00000068450
|
Spag16
|
sperm associated antigen 16 |
| chr5_-_62186372 | 5.29 |
ENSRNOT00000089789
|
Coro2a
|
coronin 2A |
| chr1_-_226353611 | 5.27 |
ENSRNOT00000037624
|
Dagla
|
diacylglycerol lipase, alpha |
| chr7_+_77763512 | 5.26 |
ENSRNOT00000006411
|
Baalc
|
brain and acute leukemia, cytoplasmic |
| chr9_-_98597359 | 5.21 |
ENSRNOT00000027506
|
Per2
|
period circadian clock 2 |
| chr6_+_10348308 | 5.19 |
ENSRNOT00000034991
|
Epas1
|
endothelial PAS domain protein 1 |
| chr14_-_14390699 | 5.17 |
ENSRNOT00000046639
|
Anxa3
|
annexin A3 |
| chr10_-_41408518 | 5.15 |
ENSRNOT00000018967
|
Nmur2
|
neuromedin U receptor 2 |
| chr12_+_49665794 | 5.15 |
ENSRNOT00000082322
|
Grk3
|
G protein-coupled receptor kinase 3 |
| chr5_-_160158386 | 5.12 |
ENSRNOT00000089345
|
Fblim1
|
filamin binding LIM protein 1 |
| chr1_-_261446570 | 5.05 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr1_+_196996581 | 5.01 |
ENSRNOT00000021690
|
Il21r
|
interleukin 21 receptor |
| chrX_+_23414354 | 5.01 |
ENSRNOT00000031235
|
Cldn34a
|
claudin 34A |
| chr1_+_88542035 | 4.98 |
ENSRNOT00000032973
|
Zfp420
|
zinc finger protein 420 |
| chr8_-_64305330 | 4.96 |
ENSRNOT00000030199
|
Tmem202
|
transmembrane protein 202 |
| chr1_-_72468738 | 4.94 |
ENSRNOT00000029136
|
Zfp628
|
zinc finger protein 628 |
| chr15_-_24199341 | 4.93 |
ENSRNOT00000015553
|
Dlgap5
|
DLG associated protein 5 |
| chr17_-_77687456 | 4.89 |
ENSRNOT00000045765
ENSRNOT00000081645 |
Frmd4a
|
FERM domain containing 4A |
| chr11_-_35697072 | 4.87 |
ENSRNOT00000039999
|
Erg
|
ERG, ETS transcription factor |
| chr1_-_101236065 | 4.82 |
ENSRNOT00000066834
|
Cd37
|
CD37 molecule |
| chr3_-_3433148 | 4.80 |
ENSRNOT00000078020
|
Camsap1
|
calmodulin regulated spectrin-associated protein 1 |
| chr2_-_202816562 | 4.78 |
ENSRNOT00000020401
|
Fam46c
|
family with sequence similarity 46, member C |
| chr1_-_242765807 | 4.77 |
ENSRNOT00000020763
|
Pgm5
|
phosphoglucomutase 5 |
| chr1_+_85380088 | 4.67 |
ENSRNOT00000090129
|
Zfp36
|
zinc finger protein 36 |
| chr2_+_80948658 | 4.67 |
ENSRNOT00000074689
|
Dnah5
|
dynein, axonemal, heavy chain 5 |
| chr8_+_118744229 | 4.67 |
ENSRNOT00000028364
|
Kif9
|
kinesin family member 9 |
| chr1_-_116567189 | 4.66 |
ENSRNOT00000048996
|
LOC100912195
|
protein BEX1-like |
| chr3_-_176465162 | 4.65 |
ENSRNOT00000048807
|
Nkain4
|
Sodium/potassium transporting ATPase interacting 4 |
| chr4_-_14490446 | 4.61 |
ENSRNOT00000009132
|
Sema3c
|
semaphorin 3C |
| chr7_-_141624972 | 4.60 |
ENSRNOT00000078785
|
AABR07058884.1
|
|
| chr13_-_85443976 | 4.56 |
ENSRNOT00000005213
|
Uck2
|
uridine-cytidine kinase 2 |
| chr9_+_81880177 | 4.54 |
ENSRNOT00000022839
|
Stk36
|
serine/threonine kinase 36 |
| chr12_-_23727535 | 4.54 |
ENSRNOT00000085911
ENSRNOT00000001950 |
Dtx2
|
deltex E3 ubiquitin ligase 2 |
| chr10_-_76039964 | 4.51 |
ENSRNOT00000003164
|
Msi2
|
musashi RNA-binding protein 2 |
| chr4_-_8255379 | 4.50 |
ENSRNOT00000076583
ENSRNOT00000076950 ENSRNOT00000076633 |
Kmt2e
|
lysine methyltransferase 2E |
| chr6_-_44363915 | 4.49 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr5_-_171710312 | 4.46 |
ENSRNOT00000076044
ENSRNOT00000071280 ENSRNOT00000072059 ENSRNOT00000077015 |
Prdm16
|
PR/SET domain 16 |
| chrX_+_70563570 | 4.43 |
ENSRNOT00000003772
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr3_-_80349145 | 4.40 |
ENSRNOT00000040633
|
RGD1309540
|
similar to hypothetical protein MGC40841; similar to hypothetical protein MGC4707 |
| chr2_-_80408672 | 4.34 |
ENSRNOT00000067638
|
Fam105a
|
family with sequence similarity 105, member A |
| chr15_+_3938075 | 4.33 |
ENSRNOT00000065644
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr5_-_159293673 | 4.24 |
ENSRNOT00000009081
|
Padi4
|
peptidyl arginine deiminase 4 |
| chr10_-_90940118 | 4.23 |
ENSRNOT00000093143
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr4_+_98481520 | 4.21 |
ENSRNOT00000078381
ENSRNOT00000048493 |
AABR07060886.1
|
|
| chr5_-_63192025 | 4.19 |
ENSRNOT00000008913
|
Alg2
|
ALG2, alpha-1,3/1,6-mannosyltransferase |
| chr20_+_7330250 | 4.16 |
ENSRNOT00000090993
|
LOC499407
|
LRRGT00097 |
| chr1_-_178196294 | 4.08 |
ENSRNOT00000067170
|
Btbd10
|
BTB domain containing 10 |
| chr4_-_27966398 | 4.06 |
ENSRNOT00000012597
|
Cdk6
|
cyclin-dependent kinase 6 |
| chr1_-_219438779 | 4.03 |
ENSRNOT00000029237
|
Tbc1d10c
|
TBC1 domain family, member 10C |
| chr16_+_19501193 | 3.96 |
ENSRNOT00000085328
|
Cyp4f18
|
cytochrome P450, family 4, subfamily f, polypeptide 18 |
| chr4_-_56114254 | 3.94 |
ENSRNOT00000010673
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr1_-_81946714 | 3.91 |
ENSRNOT00000027578
|
Grik5
|
glutamate ionotropic receptor kainate type subunit 5 |
| chr6_+_96479430 | 3.90 |
ENSRNOT00000006729
|
Prkch
|
protein kinase C, eta |
| chr3_+_5624506 | 3.89 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr19_+_39063998 | 3.85 |
ENSRNOT00000081116
|
Has3
|
hyaluronan synthase 3 |
| chr13_+_100980574 | 3.83 |
ENSRNOT00000067005
ENSRNOT00000004649 |
Capn8
|
calpain 8 |
| chr3_+_122928964 | 3.82 |
ENSRNOT00000074724
|
LOC100365450
|
rCG26795-like |
| chr1_-_72941869 | 3.80 |
ENSRNOT00000032306
|
Eps8l1
|
EPS8-like 1 |
| chr1_+_89008117 | 3.76 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chrX_-_2116656 | 3.74 |
ENSRNOT00000081913
|
Rp2
|
retinitis pigmentosa 2 (X-linked recessive) |
| chrX_+_134742356 | 3.71 |
ENSRNOT00000005267
ENSRNOT00000082363 |
Ocrl
|
OCRL, inositol polyphosphate-5-phosphatase |
| chr7_+_145117951 | 3.70 |
ENSRNOT00000055272
|
Pde1b
|
phosphodiesterase 1B |
| chr3_+_153580861 | 3.69 |
ENSRNOT00000080516
ENSRNOT00000012739 |
Src
|
SRC proto-oncogene, non-receptor tyrosine kinase |
| chrX_-_118615798 | 3.67 |
ENSRNOT00000045463
|
Lrch2
|
leucine rich repeats and calponin homology domain containing 2 |
| chr1_+_198284473 | 3.65 |
ENSRNOT00000027022
|
Doc2a
|
double C2 domain alpha |
| chr18_-_25314047 | 3.62 |
ENSRNOT00000079778
|
AABR07031674.4
|
|
| chr10_+_53466870 | 3.60 |
ENSRNOT00000057503
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr17_+_9109731 | 3.56 |
ENSRNOT00000016009
|
Cxcl14
|
C-X-C motif chemokine ligand 14 |
| chrX_+_106306795 | 3.51 |
ENSRNOT00000073661
|
Gprasp1
|
G protein-coupled receptor associated sorting protein 1 |
| chr1_-_265573117 | 3.51 |
ENSRNOT00000044195
ENSRNOT00000055915 |
LOC100911951
|
Kv channel-interacting protein 2-like |
| chr10_-_47857326 | 3.50 |
ENSRNOT00000085703
ENSRNOT00000091963 |
Epn2
|
epsin 2 |
| chr6_-_21880003 | 3.49 |
ENSRNOT00000006505
|
Ttc27
|
tetratricopeptide repeat domain 27 |
| chr6_-_3794489 | 3.47 |
ENSRNOT00000087891
ENSRNOT00000011000 ENSRNOT00000084499 |
Thumpd2
|
THUMP domain containing 2 |
| chr13_-_85443727 | 3.47 |
ENSRNOT00000090668
ENSRNOT00000076125 |
Uck2
|
uridine-cytidine kinase 2 |
| chr4_-_116278615 | 3.47 |
ENSRNOT00000020505
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr17_+_72240802 | 3.43 |
ENSRNOT00000025993
ENSRNOT00000090453 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr1_-_206394346 | 3.38 |
ENSRNOT00000038122
|
LOC100302465
|
hypothetical LOC100302465 |
| chr4_+_70572942 | 3.35 |
ENSRNOT00000051964
|
AC142181.1
|
|
| chr19_-_10358695 | 3.32 |
ENSRNOT00000019770
|
Katnb1
|
katanin regulatory subunit B1 |
| chr20_+_7718282 | 3.24 |
ENSRNOT00000000593
|
Scube3
|
signal peptide, CUB domain and EGF like domain containing 3 |
| chr5_+_144031402 | 3.23 |
ENSRNOT00000011694
|
Csf3r
|
colony stimulating factor 3 receptor |
| chr2_+_29410579 | 3.22 |
ENSRNOT00000023180
|
Zfp366
|
zinc finger protein 366 |
| chrX_-_2116483 | 3.21 |
ENSRNOT00000055077
|
Rp2
|
retinitis pigmentosa 2 (X-linked recessive) |
| chr1_-_78212350 | 3.21 |
ENSRNOT00000071098
|
Inafm1
|
InaF-motif containing 1 |
| chr6_-_6842758 | 3.20 |
ENSRNOT00000006094
|
Kcng3
|
potassium voltage-gated channel modifier subfamily G member 3 |
| chr2_+_51672722 | 3.16 |
ENSRNOT00000016485
|
Fgf10
|
fibroblast growth factor 10 |
| chr1_+_85437780 | 3.14 |
ENSRNOT00000093740
|
Supt5h
|
SPT5 homolog, DSIF elongation factor subunit |
| chr14_-_37871051 | 3.12 |
ENSRNOT00000003087
|
Slain2
|
SLAIN motif family, member 2 |
| chr16_+_9772688 | 3.12 |
ENSRNOT00000048001
|
AABR07024637.1
|
|
| chr9_+_16747662 | 3.10 |
ENSRNOT00000080815
|
Cul9
|
cullin 9 |
| chr16_+_17611115 | 3.05 |
ENSRNOT00000032309
|
Sh2d4b
|
SH2 domain containing 4B |
| chr1_-_241046249 | 3.05 |
ENSRNOT00000067796
ENSRNOT00000081866 |
Smc5
|
structural maintenance of chromosomes 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.0 | 29.9 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 8.5 | 25.5 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 7.7 | 30.9 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 7.5 | 22.5 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 7.0 | 21.0 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 5.5 | 16.4 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 5.4 | 27.2 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 5.1 | 20.5 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 5.0 | 20.0 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 2.8 | 8.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 2.7 | 8.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 2.7 | 8.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 2.6 | 7.7 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 2.5 | 7.6 | GO:0030091 | protein repair(GO:0030091) |
| 2.5 | 14.9 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 2.5 | 7.4 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 2.4 | 14.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 2.3 | 7.0 | GO:1904700 | granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 2.3 | 11.6 | GO:2000173 | insulin processing(GO:0030070) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 2.3 | 6.9 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) |
| 2.3 | 9.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 2.2 | 13.4 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 2.2 | 28.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 2.0 | 8.0 | GO:0044211 | CTP salvage(GO:0044211) |
| 2.0 | 6.0 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 2.0 | 6.0 | GO:0072237 | metanephric proximal tubule development(GO:0072237) |
| 2.0 | 4.0 | GO:0036101 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 1.9 | 14.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.8 | 5.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.8 | 5.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 1.7 | 5.2 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 1.7 | 6.9 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 1.7 | 5.1 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 1.7 | 5.1 | GO:0043474 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 1.7 | 5.1 | GO:2000040 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.7 | 18.2 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 1.5 | 4.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 1.3 | 7.6 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 1.3 | 2.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.3 | 20.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 1.2 | 3.7 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 1.2 | 6.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 1.2 | 4.8 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 1.2 | 10.7 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 1.2 | 8.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.2 | 4.7 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 1.2 | 3.5 | GO:1902809 | cornification(GO:0070268) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 1.2 | 4.6 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 1.1 | 4.5 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 1.1 | 9.0 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 1.1 | 3.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 1.0 | 5.2 | GO:0048625 | myoblast fate commitment(GO:0048625) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 1.0 | 11.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.0 | 13.0 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 1.0 | 3.9 | GO:0036118 | hyaluranon cable assembly(GO:0036118) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.9 | 13.3 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.8 | 4.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.8 | 4.9 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.8 | 7.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.8 | 6.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.8 | 3.9 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.8 | 7.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.8 | 6.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.8 | 82.3 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.7 | 2.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.7 | 7.6 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.7 | 6.8 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.6 | 4.8 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.6 | 14.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.6 | 4.1 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.6 | 5.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.6 | 4.0 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.6 | 1.7 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.5 | 3.1 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.5 | 2.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.5 | 1.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.5 | 3.9 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.5 | 2.4 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.5 | 1.9 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) histone H3-S28 phosphorylation(GO:0043988) |
| 0.5 | 5.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.4 | 2.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.4 | 6.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.4 | 3.9 | GO:0034351 | negative regulation of glial cell apoptotic process(GO:0034351) positive regulation of keratinocyte differentiation(GO:0045618) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.4 | 5.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.4 | 3.0 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.4 | 4.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.4 | 3.7 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.4 | 5.7 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.4 | 7.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.4 | 4.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 2.8 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.4 | 4.3 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.4 | 5.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.4 | 2.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.4 | 1.4 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) detection of muscle stretch(GO:0035995) |
| 0.3 | 3.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.3 | 4.7 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.3 | 4.0 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.3 | 5.2 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.3 | 1.8 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.3 | 3.5 | GO:1901679 | nucleotide transmembrane transport(GO:1901679) |
| 0.3 | 4.5 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.3 | 6.7 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.3 | 3.9 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.3 | 1.8 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 8.4 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.2 | 1.9 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.2 | 1.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 1.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 1.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 2.9 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.2 | 3.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.2 | 6.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.2 | 2.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 1.6 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.2 | 10.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.2 | 33.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.2 | 0.7 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.2 | 4.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.2 | 0.7 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.2 | 0.7 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 0.2 | 1.6 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.2 | 7.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 3.0 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.6 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.1 | 3.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 3.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 1.2 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 1.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.8 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 3.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 7.2 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 3.6 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 5.3 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.1 | 3.2 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 1.6 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 4.7 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.1 | 7.6 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 8.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 3.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 1.6 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 1.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 3.5 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.4 | GO:0019046 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.1 | 1.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.8 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.9 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.7 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 3.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 4.2 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.1 | 2.1 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 1.6 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.1 | 3.8 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.8 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 2.3 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 0.5 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 | 0.9 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 1.8 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 1.0 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 2.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 7.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 8.0 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 4.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 2.5 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.5 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 1.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 4.5 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 1.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 3.1 | GO:0007088 | regulation of mitotic nuclear division(GO:0007088) |
| 0.0 | 2.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.2 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 1.0 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 3.6 | GO:0045786 | negative regulation of cell cycle(GO:0045786) |
| 0.0 | 0.2 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 2.4 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 19.9 | GO:0032173 | septin collar(GO:0032173) |
| 5.6 | 16.7 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 3.3 | 20.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 2.1 | 8.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 1.9 | 7.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 1.7 | 18.2 | GO:0002177 | manchette(GO:0002177) |
| 1.6 | 27.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 1.5 | 6.0 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 1.3 | 9.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 1.3 | 3.9 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 1.2 | 19.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 1.1 | 5.3 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 1.0 | 13.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 1.0 | 7.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.9 | 25.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.8 | 22.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.8 | 4.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.8 | 7.0 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.8 | 6.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.7 | 11.9 | GO:0042581 | specific granule(GO:0042581) |
| 0.7 | 11.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.7 | 3.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.6 | 3.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.6 | 14.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.6 | 4.7 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.5 | 6.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.5 | 2.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.4 | 1.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.4 | 3.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.4 | 2.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 4.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 6.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.3 | 4.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.3 | 8.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 4.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.3 | 39.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.3 | 5.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.3 | 5.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 14.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.3 | 5.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.3 | 4.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 3.5 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.2 | 6.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 31.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 2.7 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.2 | 5.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.2 | 6.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 0.7 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.2 | 4.7 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 11.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 0.8 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 5.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 4.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 10.8 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 8.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 3.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 9.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 14.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 4.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 4.5 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 2.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 0.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 28.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 12.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 9.3 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 1.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 6.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 4.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 8.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 4.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 2.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 8.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 2.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 10.1 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 1.1 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.0 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.8 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 5.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.8 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 4.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.6 | GO:0030018 | Z disc(GO:0030018) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 85.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 6.2 | 30.9 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 5.0 | 14.9 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 4.9 | 14.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 4.5 | 27.2 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 3.6 | 10.7 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 3.4 | 13.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 3.2 | 13.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 3.1 | 18.8 | GO:0005499 | vitamin D binding(GO:0005499) |
| 2.4 | 22.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 2.2 | 20.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 2.1 | 8.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 2.1 | 6.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 2.0 | 4.0 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 1.9 | 7.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 1.8 | 18.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 1.8 | 5.3 | GO:0070905 | serine binding(GO:0070905) |
| 1.6 | 8.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 1.5 | 6.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.4 | 7.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 1.4 | 16.8 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 1.4 | 4.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 1.4 | 9.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.4 | 13.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 1.2 | 6.0 | GO:0015168 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 1.2 | 8.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 1.2 | 8.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.2 | 3.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 1.1 | 8.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 1.1 | 7.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 1.1 | 3.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.1 | 4.2 | GO:0034618 | arginine binding(GO:0034618) |
| 1.0 | 5.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.0 | 22.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 1.0 | 14.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.0 | 3.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.0 | 4.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.9 | 3.7 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.9 | 5.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.8 | 2.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.8 | 8.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.8 | 16.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.8 | 3.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.8 | 2.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.7 | 4.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.6 | 3.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.6 | 6.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.6 | 13.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.6 | 9.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.6 | 3.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.5 | 3.8 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.5 | 3.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 5.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.5 | 3.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.5 | 16.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.5 | 11.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.5 | 12.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.5 | 14.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.5 | 3.7 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.5 | 6.9 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.5 | 2.7 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.4 | 1.7 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.4 | 7.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.4 | 4.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.4 | 2.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.4 | 1.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.3 | 8.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 22.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 2.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 2.5 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 12.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 1.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 9.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 6.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 0.7 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.2 | 3.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 0.7 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 5.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 2.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 20.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.2 | 4.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 6.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 5.1 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.2 | 5.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.2 | 3.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 3.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 1.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 5.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 3.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 3.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 4.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 4.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.0 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 1.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 2.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.7 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 1.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 2.4 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 5.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 4.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 7.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 6.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 3.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 7.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 3.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.9 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 7.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 2.6 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 11.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 3.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 3.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 1.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 5.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 4.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 2.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 2.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 4.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 13.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 11.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 8.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 1.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.6 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.9 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 29.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.8 | 17.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.7 | 12.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.6 | 49.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.5 | 15.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.4 | 6.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.4 | 6.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 12.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 8.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 12.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.2 | 5.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 4.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 30.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 2.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 5.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 3.9 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.2 | 14.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 4.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 9.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 4.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 34.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 4.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 12.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.8 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 72.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 2.3 | 9.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 2.3 | 78.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 1.4 | 25.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 1.2 | 17.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 1.0 | 20.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.8 | 12.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.8 | 22.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.8 | 9.0 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.7 | 2.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.6 | 21.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.6 | 10.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.6 | 6.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.6 | 15.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.6 | 5.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.6 | 7.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.6 | 11.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.5 | 11.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.5 | 6.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 7.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.3 | 16.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 7.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 5.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 9.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 5.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.3 | 16.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.3 | 4.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 8.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 4.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 29.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 4.7 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.2 | 3.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 11.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 3.7 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.2 | 3.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 2.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 3.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 3.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.2 | 4.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 6.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 4.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 3.0 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 4.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 8.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.1 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 1.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 3.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.9 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 5.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |