Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
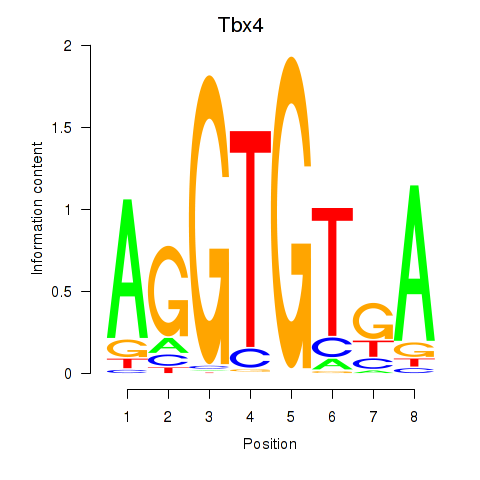
Results for Tbx4
Z-value: 0.71
Transcription factors associated with Tbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx4
|
ENSRNOG00000003544 | T-box 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx4 | rn6_v1_chr10_+_73333119_73333119 | -0.09 | 1.2e-01 | Click! |
Activity profile of Tbx4 motif
Sorted Z-values of Tbx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_174013288 | 43.04 |
ENSRNOT00000013904
|
Serpini1
|
serpin family I member 1 |
| chr11_-_4397361 | 39.92 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr5_+_143500441 | 36.94 |
ENSRNOT00000045513
|
Grik3
|
glutamate ionotropic receptor kainate type subunit 3 |
| chr4_-_52350624 | 33.30 |
ENSRNOT00000060476
|
Tmem229a
|
transmembrane protein 229A |
| chr2_+_74360622 | 33.06 |
ENSRNOT00000014013
|
Cdh18
|
cadherin 18 |
| chr7_+_54980120 | 30.05 |
ENSRNOT00000005690
ENSRNOT00000005773 |
Kcnc2
|
potassium voltage-gated channel subfamily C member 2 |
| chr11_-_59110562 | 26.48 |
ENSRNOT00000047907
ENSRNOT00000042024 |
Lsamp
|
limbic system-associated membrane protein |
| chr5_-_12563429 | 21.57 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr7_+_77066955 | 21.46 |
ENSRNOT00000008700
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr10_-_68926264 | 21.41 |
ENSRNOT00000008661
|
Phb-ps1
|
prohibitin, pseudogene 1 |
| chr5_+_164808323 | 21.40 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr9_+_81816872 | 18.09 |
ENSRNOT00000041407
|
Plcd4
|
phospholipase C, delta 4 |
| chr9_-_61810417 | 17.77 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr18_-_40134504 | 14.75 |
ENSRNOT00000022294
|
Trim36
|
tripartite motif-containing 36 |
| chr1_-_165680176 | 14.62 |
ENSRNOT00000025245
ENSRNOT00000082697 |
Plekhb1
|
pleckstrin homology domain containing B1 |
| chr3_+_131351587 | 14.19 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr4_+_71675383 | 13.67 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr5_+_136683592 | 13.11 |
ENSRNOT00000085527
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr18_+_46148849 | 11.25 |
ENSRNOT00000026724
|
Prr16
|
proline rich 16 |
| chr14_+_12218553 | 10.84 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr1_+_187149453 | 9.17 |
ENSRNOT00000082738
|
Xylt1
|
xylosyltransferase 1 |
| chr16_+_46731403 | 8.74 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr4_-_56114254 | 8.24 |
ENSRNOT00000010673
|
Lrrc4
|
leucine rich repeat containing 4 |
| chrX_+_70563570 | 8.19 |
ENSRNOT00000003772
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr4_-_83137527 | 7.66 |
ENSRNOT00000039580
|
Jazf1
|
JAZF zinc finger 1 |
| chr4_+_174181644 | 7.10 |
ENSRNOT00000011555
|
Capza3
|
capping actin protein of muscle Z-line alpha subunit 3 |
| chr6_-_50923510 | 7.07 |
ENSRNOT00000010631
|
Bcap29
|
B-cell receptor-associated protein 29 |
| chr10_+_85608544 | 7.07 |
ENSRNOT00000092122
|
Mllt6
|
MLLT6, PHD finger domain containing |
| chr13_-_55878094 | 6.68 |
ENSRNOT00000014218
|
Lhx9
|
LIM homeobox 9 |
| chr1_+_88542035 | 5.99 |
ENSRNOT00000032973
|
Zfp420
|
zinc finger protein 420 |
| chr1_-_170471076 | 5.81 |
ENSRNOT00000025159
|
Arfip2
|
ADP-ribosylation factor interacting protein 2 |
| chr16_+_32457521 | 5.44 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr3_+_100768637 | 5.33 |
ENSRNOT00000083542
|
Bdnf
|
brain-derived neurotrophic factor |
| chr18_+_53609214 | 5.26 |
ENSRNOT00000077443
|
Slc27a6
|
solute carrier family 27 member 6 |
| chr9_-_63637677 | 5.25 |
ENSRNOT00000049259
|
Satb2
|
SATB homeobox 2 |
| chr13_-_85443976 | 4.97 |
ENSRNOT00000005213
|
Uck2
|
uridine-cytidine kinase 2 |
| chr1_+_85485289 | 4.91 |
ENSRNOT00000026182
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr9_-_15700235 | 4.79 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr1_+_170471238 | 4.53 |
ENSRNOT00000076961
ENSRNOT00000075597 ENSRNOT00000076631 ENSRNOT00000076783 |
Timm10b
Dnhd1
|
translocase of inner mitochondrial membrane 10B dynein heavy chain domain 1 |
| chr9_-_49950093 | 4.22 |
ENSRNOT00000023014
|
Fhl2
|
four and a half LIM domains 2 |
| chr5_+_24434872 | 4.14 |
ENSRNOT00000010696
|
Ccne2
|
cyclin E2 |
| chr13_-_85443727 | 3.85 |
ENSRNOT00000090668
ENSRNOT00000076125 |
Uck2
|
uridine-cytidine kinase 2 |
| chr1_-_143398093 | 2.55 |
ENSRNOT00000078916
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr8_+_77640222 | 2.48 |
ENSRNOT00000079115
|
Aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr1_-_22404002 | 2.47 |
ENSRNOT00000044098
|
Taar8a
|
trace amine-associated receptor 8a |
| chr9_+_47281961 | 2.24 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr12_+_52359310 | 1.80 |
ENSRNOT00000071857
ENSRNOT00000065893 |
Fbrsl1
|
fibrosin-like 1 |
| chr1_+_185863043 | 1.77 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr20_+_2058197 | 1.72 |
ENSRNOT00000077774
ENSRNOT00000040634 |
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr4_-_100783016 | 1.57 |
ENSRNOT00000020671
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr2_-_62634785 | 0.98 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr1_-_141533908 | 0.62 |
ENSRNOT00000020117
|
Mesp1
|
mesoderm posterior bHLH transcription factor 1 |
| chr10_-_88645364 | 0.56 |
ENSRNOT00000030344
|
Rab5c
|
RAB5C, member RAS oncogene family |
| chr1_+_172822544 | 0.52 |
ENSRNOT00000090018
|
AABR07004960.1
|
|
| chr15_+_8739589 | 0.48 |
ENSRNOT00000071751
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr1_-_172322795 | 0.25 |
ENSRNOT00000075318
|
RGD1562400
|
similar to olfactory receptor MOR204-14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 21.4 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 5.0 | 30.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 3.6 | 10.8 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 3.4 | 36.9 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 3.0 | 17.8 | GO:0033227 | dsRNA transport(GO:0033227) |
| 2.4 | 7.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 2.2 | 6.7 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 2.2 | 8.8 | GO:0044211 | CTP salvage(GO:0044211) |
| 1.4 | 5.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 1.2 | 4.9 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.2 | 13.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 1.2 | 7.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 1.1 | 26.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 1.1 | 13.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.8 | 8.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.8 | 32.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.8 | 8.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.8 | 9.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.7 | 8.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.7 | 5.3 | GO:0061193 | taste bud development(GO:0061193) |
| 0.7 | 39.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.5 | 2.5 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.5 | 5.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.5 | 11.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.4 | 1.8 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.3 | 5.8 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.3 | 7.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.3 | 4.2 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.3 | 33.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 0.6 | GO:0060974 | growth involved in heart morphogenesis(GO:0003241) cell migration involved in heart formation(GO:0060974) regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.2 | 2.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 43.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 4.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 14.2 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 0.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 5.3 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 17.7 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 1.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.5 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.0 | 4.5 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 10.6 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 70.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 1.1 | 21.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 1.1 | 36.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.9 | 4.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.8 | 21.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.7 | 7.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 5.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.3 | 5.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 13.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 26.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 5.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 14.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 4.2 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 10.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 7.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 14.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 22.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 8.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 17.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 8.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 13.1 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 35.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 9.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 88.4 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 36.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 3.1 | 9.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 2.2 | 10.8 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 1.8 | 5.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.8 | 8.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 1.4 | 8.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.3 | 30.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 1.0 | 13.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) glycine transmembrane transporter activity(GO:0015187) |
| 0.9 | 21.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.8 | 18.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.8 | 13.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.7 | 5.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.5 | 5.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.4 | 43.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 2.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 4.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 5.8 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 2.5 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 2.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 14.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 39.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 4.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 14.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 33.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 1.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 8.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 41.3 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 5.9 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 17.8 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 7.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.6 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 4.8 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 8.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 39.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 4.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 21.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 5.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 5.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 73.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 2.3 | 36.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.2 | 10.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.8 | 21.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.6 | 30.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.6 | 13.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 8.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 5.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 4.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 14.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 4.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |