Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
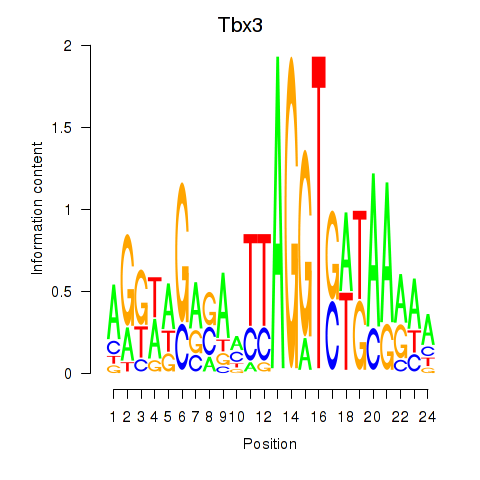
Results for Tbx3
Z-value: 0.56
Transcription factors associated with Tbx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx3
|
ENSRNOG00000008706 | T-box 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx3 | rn6_v1_chr12_-_42492526_42492526 | 0.09 | 1.0e-01 | Click! |
Activity profile of Tbx3 motif
Sorted Z-values of Tbx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_80166807 | 23.75 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr1_+_222310920 | 23.55 |
ENSRNOT00000091465
|
Macrod1
|
MACRO domain containing 1 |
| chr9_-_80167033 | 23.22 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr1_+_72882806 | 21.51 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr13_-_51297621 | 13.04 |
ENSRNOT00000030926
|
Ndufv3
|
NADH:ubiquinone oxidoreductase subunit V3 |
| chr2_-_30748325 | 12.93 |
ENSRNOT00000084294
ENSRNOT00000083089 |
Mrps36
|
mitochondrial ribosomal protein S36 |
| chr17_+_69468427 | 10.98 |
ENSRNOT00000058413
|
RGD1564865
|
similar to 20-alpha-hydroxysteroid dehydrogenase |
| chr4_+_30313102 | 10.77 |
ENSRNOT00000012657
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr20_-_14193690 | 10.04 |
ENSRNOT00000058237
|
Upb1
|
beta-ureidopropionase 1 |
| chr5_-_171710312 | 8.44 |
ENSRNOT00000076044
ENSRNOT00000071280 ENSRNOT00000072059 ENSRNOT00000077015 |
Prdm16
|
PR/SET domain 16 |
| chr6_-_108076186 | 8.39 |
ENSRNOT00000014814
|
Fam161b
|
family with sequence similarity 161, member B |
| chr8_-_7426611 | 7.17 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr6_+_108076306 | 6.73 |
ENSRNOT00000014913
|
Coq6
|
coenzyme Q6 monooxygenase |
| chr13_-_36290531 | 5.00 |
ENSRNOT00000071388
|
Steap3
|
STEAP3 metalloreductase |
| chr1_+_56288832 | 4.84 |
ENSRNOT00000079718
|
Smoc2
|
SPARC related modular calcium binding 2 |
| chr20_+_37876650 | 4.83 |
ENSRNOT00000001054
|
Gja1
|
gap junction protein, alpha 1 |
| chr3_-_120373500 | 4.65 |
ENSRNOT00000067727
|
Nphp1
|
nephrocystin 1 |
| chr7_-_29701586 | 4.62 |
ENSRNOT00000009084
ENSRNOT00000089269 |
Ano4
|
anoctamin 4 |
| chr1_+_185673177 | 4.52 |
ENSRNOT00000048020
|
Sox6
|
SRY box 6 |
| chr3_-_146396299 | 3.29 |
ENSRNOT00000040188
ENSRNOT00000008931 |
Apmap
|
adipocyte plasma membrane associated protein |
| chr4_+_72292261 | 2.96 |
ENSRNOT00000078723
|
LOC103690255
|
olfactory receptor-like protein OLF3 |
| chr1_-_188097530 | 2.85 |
ENSRNOT00000078825
|
Syt17
|
synaptotagmin 17 |
| chr9_-_79530821 | 2.68 |
ENSRNOT00000085619
|
Mreg
|
melanoregulin |
| chr2_+_189922996 | 2.03 |
ENSRNOT00000016499
|
S100a16
|
S100 calcium binding protein A16 |
| chr4_-_161658519 | 2.02 |
ENSRNOT00000007634
ENSRNOT00000067895 |
Tulp3
|
tubby-like protein 3 |
| chr16_+_70644474 | 1.96 |
ENSRNOT00000045955
|
LOC100359503
|
ribosomal protein S28-like |
| chr17_+_38323674 | 1.23 |
ENSRNOT00000021982
|
Prl5a1
|
prolactin family 5, subfamily a, member 1 |
| chr10_-_60772313 | 1.18 |
ENSRNOT00000050847
|
Olr1504
|
olfactory receptor 1504 |
| chr7_-_70355619 | 1.16 |
ENSRNOT00000031272
|
Tspan31
|
tetraspanin 31 |
| chr1_-_188097374 | 0.95 |
ENSRNOT00000092246
|
Syt17
|
synaptotagmin 17 |
| chrX_+_97074710 | 0.71 |
ENSRNOT00000044379
|
RGD1561230
|
similar to RIKEN cDNA 4921511C20 gene |
| chr10_-_44256945 | 0.68 |
ENSRNOT00000003818
|
Olr1432
|
olfactory receptor 1432 |
| chr3_+_122928964 | 0.66 |
ENSRNOT00000074724
|
LOC100365450
|
rCG26795-like |
| chr5_+_137255924 | 0.56 |
ENSRNOT00000088022
ENSRNOT00000092755 |
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr3_-_18244535 | 0.52 |
ENSRNOT00000040689
|
AABR07051626.1
|
|
| chr2_+_188561429 | 0.39 |
ENSRNOT00000076458
ENSRNOT00000027867 |
Krtcap2
|
keratinocyte associated protein 2 |
| chr3_-_149563476 | 0.26 |
ENSRNOT00000071869
|
AABR07054352.2
|
|
| chr1_+_222746023 | 0.21 |
ENSRNOT00000028787
|
Atl3
|
atlastin GTPase 3 |
| chr17_+_2690062 | 0.20 |
ENSRNOT00000024937
|
Olr1653
|
olfactory receptor 1653 |
| chr2_+_4252496 | 0.20 |
ENSRNOT00000071535
|
RGD1560883
|
similar to KIAA0825 protein |
| chr19_+_10142496 | 0.18 |
ENSRNOT00000088645
ENSRNOT00000060351 |
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr8_-_130127392 | 0.14 |
ENSRNOT00000026159
|
Cck
|
cholecystokinin |
| chr17_-_42737000 | 0.08 |
ENSRNOT00000077923
|
Prl3a1
|
Prolactin family 3, subfamily a, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.7 | 47.0 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 5.9 | 23.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 2.5 | 10.0 | GO:0019483 | beta-alanine biosynthetic process(GO:0019483) |
| 2.4 | 7.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 1.7 | 5.0 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 1.6 | 4.8 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 1.5 | 4.6 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 1.4 | 21.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.9 | 8.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.9 | 4.6 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.8 | 10.8 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.8 | 4.5 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.4 | 6.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.4 | 2.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 13.0 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 2.7 | GO:0032401 | establishment of melanosome localization(GO:0032401) melanosome transport(GO:0032402) |
| 0.1 | 3.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.6 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 2.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 4.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.4 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.1 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 2.0 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 11.0 | GO:0055114 | oxidation-reduction process(GO:0055114) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 47.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 5.4 | 21.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 2.6 | 12.9 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) oxoglutarate dehydrogenase complex(GO:0045252) |
| 1.3 | 10.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 2.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.3 | 4.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 13.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 4.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 8.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 4.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 5.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 2.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 3.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 21.9 | GO:0005739 | mitochondrion(GO:0005739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 21.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 5.2 | 47.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.2 | 5.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 1.0 | 4.8 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.5 | 23.5 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.5 | 3.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 6.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 10.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.2 | 4.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 8.4 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 0.6 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 3.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 10.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 4.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 4.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 7.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 11.0 | GO:0016491 | oxidoreductase activity(GO:0016491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 51.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 13.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 5.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 47.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.8 | 10.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.6 | 21.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 4.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 13.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 5.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 10.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |