Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
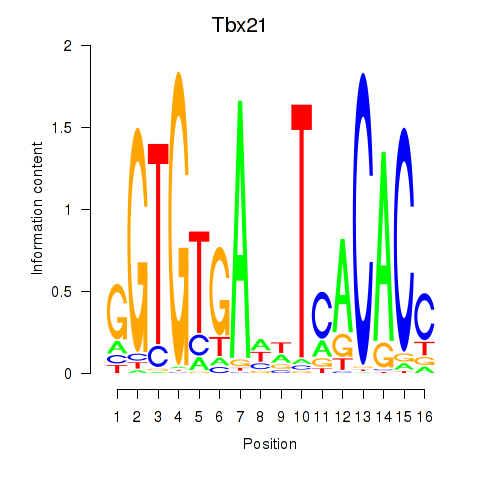
Results for Tbx21
Z-value: 1.37
Transcription factors associated with Tbx21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx21
|
ENSRNOG00000009427 | T-box 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx21 | rn6_v1_chr10_-_85049331_85049331 | 0.37 | 1.0e-11 | Click! |
Activity profile of Tbx21 motif
Sorted Z-values of Tbx21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_138239306 | 39.24 |
ENSRNOT00000039305
|
Ermap
|
erythroblast membrane-associated protein |
| chr13_+_82369493 | 38.13 |
ENSRNOT00000003733
|
Sell
|
selectin L |
| chr6_-_138550576 | 37.49 |
ENSRNOT00000075284
|
AABR07065645.1
|
|
| chr6_-_138744480 | 35.49 |
ENSRNOT00000089387
|
AABR07065651.5
|
|
| chr14_+_38030189 | 33.09 |
ENSRNOT00000035623
|
Txk
|
TXK tyrosine kinase |
| chr6_-_138909105 | 26.90 |
ENSRNOT00000087855
|
AABR07065656.9
|
|
| chr7_+_140758615 | 26.84 |
ENSRNOT00000089448
|
Troap
|
trophinin associated protein |
| chr1_+_199495298 | 26.72 |
ENSRNOT00000086003
ENSRNOT00000026748 |
Itgad
|
integrin subunit alpha D |
| chr8_-_36760742 | 24.76 |
ENSRNOT00000017307
|
Ddx25
|
DEAD-box helicase 25 |
| chr6_-_28967902 | 24.64 |
ENSRNOT00000032664
|
Mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr6_-_138772894 | 24.20 |
ENSRNOT00000080779
|
AABR07065651.1
|
|
| chr3_+_17180411 | 24.20 |
ENSRNOT00000058260
|
AABR07051565.1
|
|
| chr1_-_198476476 | 24.07 |
ENSRNOT00000027525
|
Kif22
|
kinesin family member 22 |
| chr6_-_138772736 | 23.96 |
ENSRNOT00000071492
|
AABR07065651.1
|
|
| chr17_-_44841382 | 23.71 |
ENSRNOT00000080119
|
Hist1h2ak
|
histone cluster 1, H2ak |
| chr5_+_152680407 | 23.60 |
ENSRNOT00000076864
|
Stmn1
|
stathmin 1 |
| chr2_+_87418517 | 22.39 |
ENSRNOT00000048046
|
LOC100909879
|
tyrosine-protein phosphatase non-receptor type substrate 1-like |
| chr19_+_52217984 | 21.76 |
ENSRNOT00000079580
|
Dnaaf1
|
dynein, axonemal, assembly factor 1 |
| chr10_+_72486152 | 21.40 |
ENSRNOT00000004193
|
RGD1310166
|
similar to Chromodomain-helicase-DNA-binding protein 1 (CHD-1) |
| chrX_+_138046494 | 21.32 |
ENSRNOT00000010596
|
Stk26
|
serine/threonine kinase 26 |
| chr1_-_20417637 | 21.24 |
ENSRNOT00000036049
|
RGD1559962
|
similar to High mobility group protein 2 (HMG-2) |
| chr18_+_86307646 | 21.02 |
ENSRNOT00000091778
|
Cd226
|
CD226 molecule |
| chr20_-_3401273 | 21.01 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr15_+_28074953 | 20.47 |
ENSRNOT00000086427
|
LOC103690354
|
ribonuclease pancreatic beta-type |
| chr20_-_11815647 | 19.99 |
ENSRNOT00000001639
|
Itgb2
|
integrin subunit beta 2 |
| chr10_+_105498728 | 19.57 |
ENSRNOT00000032163
|
Sphk1
|
sphingosine kinase 1 |
| chr6_+_132551394 | 19.35 |
ENSRNOT00000019583
|
Evl
|
Enah/Vasp-like |
| chr8_-_62410338 | 19.11 |
ENSRNOT00000026358
|
Csk
|
c-src tyrosine kinase |
| chr6_+_139405966 | 19.04 |
ENSRNOT00000088974
|
AABR07065693.3
|
|
| chr10_-_110232843 | 18.84 |
ENSRNOT00000054934
|
Cd7
|
Cd7 molecule |
| chr5_+_122508388 | 18.53 |
ENSRNOT00000038410
|
Tctex1d1
|
Tctex1 domain containing 1 |
| chr6_+_139523337 | 18.26 |
ENSRNOT00000090711
|
AABR07065699.4
|
|
| chr6_+_139523495 | 17.85 |
ENSRNOT00000075467
|
AABR07065699.4
|
|
| chr6_-_138852571 | 17.74 |
ENSRNOT00000081803
|
AABR07065656.8
|
|
| chr1_-_100234040 | 17.71 |
ENSRNOT00000079199
|
Acpt
|
acid phosphatase, testicular |
| chr10_+_91254058 | 17.41 |
ENSRNOT00000087218
ENSRNOT00000065373 |
Fmnl1
|
formin-like 1 |
| chr10_-_104054805 | 17.33 |
ENSRNOT00000004853
|
Nt5c
|
5', 3'-nucleotidase, cytosolic |
| chr12_+_14021727 | 16.69 |
ENSRNOT00000060608
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr7_+_119820537 | 16.53 |
ENSRNOT00000077256
ENSRNOT00000056221 |
Cyth4
|
cytohesin 4 |
| chr6_-_138685656 | 16.28 |
ENSRNOT00000041706
|
AABR07065651.7
|
|
| chr17_+_44520537 | 15.71 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr1_+_63734135 | 15.57 |
ENSRNOT00000084439
|
Nilr1
|
neutrophil immunoglobulin-like receptor-1 |
| chr5_-_122642202 | 15.33 |
ENSRNOT00000046691
|
Wdr78
|
WD repeat domain 78 |
| chr15_+_30270740 | 15.22 |
ENSRNOT00000070991
|
AABR07017707.1
|
|
| chr1_+_201672528 | 15.17 |
ENSRNOT00000093490
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr4_-_164406146 | 14.90 |
ENSRNOT00000090110
|
Klra22
|
killer cell lectin-like receptor subfamily A, member 22 |
| chr6_-_138507294 | 14.81 |
ENSRNOT00000078516
|
AABR07065640.1
|
|
| chr2_+_209661244 | 14.72 |
ENSRNOT00000091973
|
AABR07012826.1
|
|
| chr2_-_200513564 | 14.45 |
ENSRNOT00000056173
|
Phgdh
|
phosphoglycerate dehydrogenase |
| chr3_-_26056818 | 14.35 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chr11_-_69355854 | 14.09 |
ENSRNOT00000002975
|
Ropn1
|
rhophilin associated tail protein 1 |
| chr18_+_62850588 | 14.05 |
ENSRNOT00000025172
|
Gnal
|
G protein subunit alpha L |
| chrX_+_156274800 | 13.99 |
ENSRNOT00000080887
|
G6pd
|
glucose-6-phosphate dehydrogenase |
| chr12_+_16913312 | 13.66 |
ENSRNOT00000001718
|
Tmem184a
|
transmembrane protein 184A |
| chr5_-_151459037 | 13.45 |
ENSRNOT00000064472
ENSRNOT00000087836 |
Sytl1
|
synaptotagmin-like 1 |
| chr5_+_69833225 | 13.34 |
ENSRNOT00000013759
|
Nipsnap3b
|
nipsnap homolog 3B |
| chr10_+_57239993 | 13.24 |
ENSRNOT00000067919
|
LOC687707
|
hypothetical protein LOC687707 |
| chr18_-_77322690 | 13.04 |
ENSRNOT00000058382
|
Nfatc1
|
nuclear factor of activated T-cells 1 |
| chr8_+_28454962 | 12.96 |
ENSRNOT00000051573
|
Spata19
|
spermatogenesis associated 19 |
| chr2_+_30664217 | 12.17 |
ENSRNOT00000076786
|
Ak6
|
adenylate kinase 6 |
| chr16_-_31301880 | 12.12 |
ENSRNOT00000084847
ENSRNOT00000083943 |
AABR07025272.1
|
|
| chr14_-_83219071 | 12.09 |
ENSRNOT00000085788
|
Depdc5
|
DEP domain containing 5 |
| chr6_-_139660666 | 12.09 |
ENSRNOT00000086098
|
AABR07065705.1
|
|
| chr9_+_14951047 | 11.99 |
ENSRNOT00000074267
|
LOC100911489
|
uncharacterized LOC100911489 |
| chr5_+_152681101 | 11.98 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr1_+_212714179 | 11.96 |
ENSRNOT00000054880
ENSRNOT00000025680 |
Olr292
|
olfactory receptor 292 |
| chr17_-_44520240 | 11.90 |
ENSRNOT00000086538
|
Hist1h2bh
|
histone cluster 1 H2B family member h |
| chr1_-_189182306 | 11.73 |
ENSRNOT00000021249
|
Gp2
|
glycoprotein 2 |
| chr3_-_113405829 | 11.69 |
ENSRNOT00000036823
|
Ell3
|
elongation factor for RNA polymerase II 3 |
| chr18_-_70924708 | 11.55 |
ENSRNOT00000025257
|
Lipg
|
lipase G, endothelial type |
| chr2_-_200762206 | 11.38 |
ENSRNOT00000068511
ENSRNOT00000086835 |
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr1_+_222250995 | 11.35 |
ENSRNOT00000031458
|
Trpt1
|
tRNA phosphotransferase 1 |
| chr10_-_87564327 | 11.28 |
ENSRNOT00000064760
ENSRNOT00000068237 |
LOC680160
|
similar to keratin associated protein 4-7 |
| chr6_-_92643847 | 11.28 |
ENSRNOT00000009183
|
Pygl
|
glycogen phosphorylase L |
| chr6_-_138550417 | 11.19 |
ENSRNOT00000071945
|
AABR07065645.1
|
|
| chrX_-_123092217 | 11.11 |
ENSRNOT00000039710
|
Gm14569
|
predicted gene 14569 |
| chr20_-_34929965 | 11.01 |
ENSRNOT00000004499
|
Mcm9
|
minichromosome maintenance 9 homologous recombination repair factor |
| chr1_-_101651668 | 10.62 |
ENSRNOT00000028519
|
Fut2
|
fucosyltransferase 2 |
| chr19_-_56731372 | 10.52 |
ENSRNOT00000024182
|
Nup133
|
nucleoporin 133 |
| chr2_-_30664163 | 10.45 |
ENSRNOT00000024801
|
Rad17
|
RAD17 checkpoint clamp loader component |
| chr8_-_62893087 | 10.38 |
ENSRNOT00000058490
|
Ccdc33
|
coiled-coil domain containing 33 |
| chr4_-_162025090 | 10.00 |
ENSRNOT00000085887
ENSRNOT00000009904 |
Klrb1a
|
killer cell lectin-like receptor subfamily B, member 1A |
| chr14_-_5859581 | 9.98 |
ENSRNOT00000052308
|
AABR07014241.1
|
|
| chr15_-_42751330 | 9.78 |
ENSRNOT00000066478
|
Adam2
|
ADAM metallopeptidase domain 2 |
| chr3_-_2841331 | 9.46 |
ENSRNOT00000061874
|
Ccdc183
|
coiled-coil domain containing 183 |
| chr4_-_164536556 | 9.44 |
ENSRNOT00000087796
|
Ly49i2
|
Ly49 inhibitory receptor 2 |
| chr10_-_14630151 | 9.43 |
ENSRNOT00000024406
|
Ube2i
|
ubiquitin-conjugating enzyme E2I |
| chr14_-_82055290 | 9.41 |
ENSRNOT00000058062
|
RGD1560394
|
RGD1560394 |
| chr6_+_86852323 | 9.28 |
ENSRNOT00000059244
|
Fancm
|
Fanconi anemia, complementation group M |
| chr4_-_122337389 | 9.13 |
ENSRNOT00000024222
|
Cfap100
|
cilia and flagella associated protein 100 |
| chr3_-_7278758 | 9.08 |
ENSRNOT00000017004
|
Spaca9
|
sperm acrosome associated 9 |
| chr2_-_189254628 | 9.03 |
ENSRNOT00000028234
|
Il6r
|
interleukin 6 receptor |
| chr1_-_67284864 | 8.94 |
ENSRNOT00000082908
|
LOC691661
|
similar to zinc finger and SCAN domain containing 4 |
| chr9_+_73418607 | 8.62 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chrX_-_106576314 | 8.54 |
ENSRNOT00000037823
|
Nxf3
|
nuclear RNA export factor 3 |
| chr17_-_10364503 | 8.52 |
ENSRNOT00000086380
ENSRNOT00000041709 |
Tspan17
|
tetraspanin 17 |
| chr4_+_123801174 | 8.46 |
ENSRNOT00000029055
|
RGD1560289
|
similar to chromosome 3 open reading frame 20 |
| chr1_-_24982606 | 8.40 |
ENSRNOT00000074143
|
RGD1562660
|
RGD1562660 |
| chr20_-_6251634 | 8.39 |
ENSRNOT00000059194
|
Stk38
|
serine/threonine kinase 38 |
| chr7_+_2458264 | 8.26 |
ENSRNOT00000003550
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr9_+_16825845 | 8.25 |
ENSRNOT00000074821
|
LOC100910410
|
ly6/PLAUR domain-containing protein 4-like |
| chr19_-_28666993 | 8.07 |
ENSRNOT00000083821
|
AABR07043453.1
|
|
| chr3_+_70327193 | 8.04 |
ENSRNOT00000089165
|
Fsip2
|
fibrous sheath-interacting protein 2 |
| chr3_-_46601409 | 7.91 |
ENSRNOT00000079261
|
Pla2r1
|
phospholipase A2 receptor 1 |
| chr2_-_198439454 | 7.78 |
ENSRNOT00000028780
|
Fcgr1a
|
Fc fragment of IgG receptor Ia |
| chr1_-_84353725 | 7.60 |
ENSRNOT00000057177
|
Pld3
|
phospholipase D family, member 3 |
| chr9_-_104700609 | 7.39 |
ENSRNOT00000081815
ENSRNOT00000018503 |
Slco6c1
|
solute carrier organic anion transporter family, member 6c1 |
| chr19_-_28751584 | 7.31 |
ENSRNOT00000079270
|
AABR07043456.1
|
|
| chr9_+_16139101 | 7.30 |
ENSRNOT00000070803
|
LOC100910668
|
uncharacterized LOC100910668 |
| chr6_+_75429729 | 7.27 |
ENSRNOT00000043250
|
Rps10l1
|
ribosomal protein S10-like 1 |
| chr19_+_37600148 | 7.21 |
ENSRNOT00000023853
|
Ctcf
|
CCCTC-binding factor |
| chr3_-_13865843 | 7.09 |
ENSRNOT00000025194
|
Rabepk
|
Rab9 effector protein with kelch motifs |
| chr2_+_209358760 | 7.08 |
ENSRNOT00000082631
|
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr19_-_42180981 | 7.00 |
ENSRNOT00000019577
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr12_-_9727489 | 7.00 |
ENSRNOT00000001261
|
Polr1d
|
RNA polymerase I subunit D |
| chr11_+_82373870 | 6.85 |
ENSRNOT00000002429
|
Tra2b
|
transformer 2 beta homolog (Drosophila) |
| chr1_+_20022793 | 6.72 |
ENSRNOT00000061404
|
L3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr13_-_74005486 | 6.39 |
ENSRNOT00000090173
|
AABR07021465.1
|
|
| chr4_+_6282278 | 6.35 |
ENSRNOT00000010349
|
Kmt2c
|
lysine methyltransferase 2C |
| chr19_-_28044531 | 6.35 |
ENSRNOT00000086160
|
AABR07043395.1
|
|
| chr20_+_48552289 | 6.35 |
ENSRNOT00000077926
|
Cdc40
|
cell division cycle 40 |
| chr6_+_43363792 | 6.31 |
ENSRNOT00000091061
|
Cpsf3
|
cleavage and polyadenylation specific factor 3 |
| chr8_+_50563047 | 6.30 |
ENSRNOT00000025149
|
Zfp259
|
zinc finger protein 259 |
| chr17_+_56109549 | 6.25 |
ENSRNOT00000022190
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr11_+_67451549 | 6.24 |
ENSRNOT00000042777
|
Stfa2l2
|
stefin A2-like 2 |
| chr4_+_118243132 | 6.23 |
ENSRNOT00000023654
|
RGD1306746
|
similar to Hypothetical protein MGC25529 |
| chr1_-_24891461 | 6.15 |
ENSRNOT00000049208
|
LOC501467
|
similar to spermatogenesis associated glutamate (E)-rich protein 4d |
| chr15_-_40545824 | 6.14 |
ENSRNOT00000017204
|
Nup58
|
nucleoporin 58 |
| chr10_-_57837602 | 6.11 |
ENSRNOT00000075185
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr5_+_164972480 | 6.01 |
ENSRNOT00000012670
|
Fbxo2
|
F-box protein 2 |
| chr7_-_11406771 | 6.00 |
ENSRNOT00000047450
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr1_-_141533908 | 5.98 |
ENSRNOT00000020117
|
Mesp1
|
mesoderm posterior bHLH transcription factor 1 |
| chr15_+_45422010 | 5.72 |
ENSRNOT00000012231
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr4_-_179700130 | 5.55 |
ENSRNOT00000021306
|
Lmntd1
|
lamin tail domain containing 1 |
| chr12_-_22478752 | 5.54 |
ENSRNOT00000089392
ENSRNOT00000086915 |
Ache
|
acetylcholinesterase |
| chr7_+_130542202 | 5.49 |
ENSRNOT00000079501
ENSRNOT00000045647 |
Acr
|
acrosin |
| chr19_+_52258947 | 5.43 |
ENSRNOT00000021072
|
Adad2
|
adenosine deaminase domain containing 2 |
| chr4_-_155867708 | 5.37 |
ENSRNOT00000051525
|
Clec4a2
|
C-type lectin domain family 4, member A2 |
| chr10_-_83655182 | 5.29 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr8_+_117660964 | 5.25 |
ENSRNOT00000049240
|
Tmem89
|
transmembrane protein 89 |
| chr1_+_196095214 | 5.22 |
ENSRNOT00000080741
|
LOC691716
|
similar to ribosomal protein S15a |
| chr10_+_109786222 | 5.20 |
ENSRNOT00000054953
|
Npb
|
neuropeptide B |
| chr16_+_69048730 | 5.16 |
ENSRNOT00000086082
ENSRNOT00000078128 |
Rab11fip1
|
RAB11 family interacting protein 1 |
| chr2_-_206274079 | 5.03 |
ENSRNOT00000056079
|
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr7_+_123531682 | 4.96 |
ENSRNOT00000010606
|
Wbp2nl
|
WBP2 N-terminal like |
| chrX_-_124870329 | 4.91 |
ENSRNOT00000065023
|
Cul4b
|
cullin 4B |
| chrX_+_74497262 | 4.89 |
ENSRNOT00000003899
|
Zcchc13
|
zinc finger CCHC-type containing 13 |
| chr4_+_171250818 | 4.83 |
ENSRNOT00000040576
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr10_-_92602082 | 4.75 |
ENSRNOT00000007963
|
Cdc27
|
cell division cycle 27 |
| chr4_-_164691405 | 4.75 |
ENSRNOT00000090979
ENSRNOT00000091932 ENSRNOT00000078219 |
Ly49s4
Ly49i2
|
Ly49 stimulatory receptor 4 Ly49 inhibitory receptor 2 |
| chr13_+_84588351 | 4.75 |
ENSRNOT00000005090
|
Tada1
|
transcriptional adaptor 1 |
| chr3_+_2547986 | 4.42 |
ENSRNOT00000016846
|
Man1b1
|
mannosidase, alpha, class 1B, member 1 |
| chr2_-_57196568 | 4.41 |
ENSRNOT00000060146
|
Wdr70
|
WD repeat domain 70 |
| chr7_+_11054333 | 4.37 |
ENSRNOT00000007202
|
Gna15
|
guanine nucleotide binding protein, alpha 15 |
| chr17_+_79749747 | 4.33 |
ENSRNOT00000081743
|
AABR07028665.1
|
|
| chr9_-_61033553 | 4.29 |
ENSRNOT00000002864
ENSRNOT00000082681 |
Gtf3c3
|
general transcription factor IIIC subunit 3 |
| chr3_+_152752091 | 4.29 |
ENSRNOT00000037177
|
Dlgap4
|
DLG associated protein 4 |
| chr10_-_103826448 | 4.28 |
ENSRNOT00000085636
|
Fdxr
|
ferredoxin reductase |
| chr9_+_38297322 | 4.05 |
ENSRNOT00000078157
ENSRNOT00000088824 |
Bend6
|
BEN domain containing 6 |
| chr19_+_248622 | 3.95 |
ENSRNOT00000078228
|
AABR07042607.1
|
|
| chr6_-_103187275 | 3.93 |
ENSRNOT00000083272
|
LOC100359616
|
ribosomal protein L36-like |
| chr7_+_73270455 | 3.87 |
ENSRNOT00000007190
|
Pop1
|
POP1 homolog, ribonuclease P/MRP subunit |
| chr9_-_81565416 | 3.85 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr3_+_71020534 | 3.84 |
ENSRNOT00000007276
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
| chr10_+_70689863 | 3.80 |
ENSRNOT00000091122
ENSRNOT00000086987 ENSRNOT00000082030 |
Taf15
|
TATA-box binding protein associated factor 15 |
| chr11_+_86852711 | 3.78 |
ENSRNOT00000002581
|
Dgcr8
|
DGCR8 microprocessor complex subunit |
| chr5_-_150704117 | 3.75 |
ENSRNOT00000067455
|
Sesn2
|
sestrin 2 |
| chr7_-_27213651 | 3.70 |
ENSRNOT00000077041
ENSRNOT00000076468 |
Tdg
|
thymine-DNA glycosylase |
| chr4_-_113764532 | 3.66 |
ENSRNOT00000009269
|
Sema4f
|
ssemaphorin 4F |
| chr4_+_87026530 | 3.65 |
ENSRNOT00000018425
|
Avl9
|
AVL9 cell migration associated |
| chr7_-_130288509 | 3.61 |
ENSRNOT00000072955
|
Sbf1
|
SET binding factor 1 |
| chr16_+_80729959 | 3.33 |
ENSRNOT00000082049
|
Tdrp
|
testis development related protein |
| chr15_+_32324830 | 3.30 |
ENSRNOT00000071598
|
AABR07017895.1
|
|
| chr16_-_86582984 | 3.13 |
ENSRNOT00000057349
|
RGD1564807
|
similar to zinc finger protein, subfamily 1A, 5 |
| chr1_-_263959318 | 3.11 |
ENSRNOT00000068007
|
Pkd2l1
|
polycystin 2 like 1, transient receptor potential cation channel |
| chr1_+_78069193 | 3.05 |
ENSRNOT00000068406
|
Meis3
|
Meis homeobox 3 |
| chr8_-_79119720 | 3.01 |
ENSRNOT00000086562
|
Tex9
|
testis expressed 9 |
| chr15_+_34926686 | 2.95 |
ENSRNOT00000045948
|
Mcpt1l3
|
mast cell protease 1-like 3 |
| chr1_+_89162639 | 2.92 |
ENSRNOT00000028508
|
Atp4a
|
ATPase H+/K+ transporting alpha subunit |
| chr10_+_71332803 | 2.88 |
ENSRNOT00000087428
|
Synrg
|
synergin, gamma |
| chr7_-_129919946 | 2.61 |
ENSRNOT00000079976
|
AABR07058658.1
|
|
| chr3_-_161819029 | 2.60 |
ENSRNOT00000091834
|
LOC102555457
|
engulfment and cell motility protein 2-like |
| chr5_-_143120165 | 2.56 |
ENSRNOT00000012314
|
Zc3h12a
|
zinc finger CCCH type containing 12A |
| chr6_-_91250138 | 2.53 |
ENSRNOT00000052408
|
LOC100911256
|
ninein-like |
| chr17_+_57040023 | 2.52 |
ENSRNOT00000020204
|
Crem
|
cAMP responsive element modulator |
| chr20_+_47596575 | 2.50 |
ENSRNOT00000087230
ENSRNOT00000064905 |
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr10_-_55997299 | 2.44 |
ENSRNOT00000074505
|
Cyb5d1
|
cytochrome b5 domain containing 1 |
| chr10_+_71008709 | 2.36 |
ENSRNOT00000055923
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr3_-_46051096 | 2.36 |
ENSRNOT00000081302
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr2_+_187990242 | 2.34 |
ENSRNOT00000092819
|
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr7_+_117643206 | 2.33 |
ENSRNOT00000077588
|
Adck5
|
aarF domain containing kinase 5 |
| chr5_-_58288125 | 2.28 |
ENSRNOT00000068752
|
Fam205a
|
family with sequence similarity 205, member A |
| chr5_-_69752314 | 2.26 |
ENSRNOT00000024724
|
Olr852
|
olfactory receptor 852 |
| chr20_+_2044200 | 2.24 |
ENSRNOT00000092626
|
RT1-M4
|
RT1 class Ib, locus M4 |
| chr10_-_109840047 | 2.16 |
ENSRNOT00000054947
|
Notum
|
NOTUM, palmitoleoyl-protein carboxylesterase |
| chr9_+_98621506 | 2.15 |
ENSRNOT00000056612
ENSRNOT00000081992 |
Traf3ip1
|
TRAF3 interacting protein 1 |
| chr3_-_125533600 | 2.07 |
ENSRNOT00000040802
|
Lrrn4
|
leucine rich repeat neuronal 4 |
| chr7_+_144503534 | 2.07 |
ENSRNOT00000021543
|
Tbca
|
tubulin folding cofactor A |
| chr20_+_48504264 | 2.06 |
ENSRNOT00000087740
|
Cdc40
|
cell division cycle 40 |
| chr5_-_151029233 | 1.96 |
ENSRNOT00000089155
|
Ppp1r8
|
protein phosphatase 1, regulatory subunit 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx21
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 35.6 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 8.3 | 33.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 7.0 | 21.0 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 4.7 | 14.0 | GO:1904879 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 4.3 | 17.3 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 4.2 | 38.1 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 3.9 | 19.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 3.8 | 11.5 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 3.8 | 19.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 3.4 | 13.7 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 3.3 | 13.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 3.1 | 21.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 3.0 | 6.0 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 3.0 | 26.7 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 2.9 | 11.7 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 2.9 | 14.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) threonine metabolic process(GO:0006566) |
| 2.8 | 11.3 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 2.8 | 8.3 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 2.6 | 7.8 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 1.9 | 11.4 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) |
| 1.8 | 9.0 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 1.8 | 21.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 1.8 | 14.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 1.6 | 19.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.6 | 6.3 | GO:0030576 | trophectodermal cell proliferation(GO:0001834) Cajal body organization(GO:0030576) |
| 1.5 | 10.5 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 1.5 | 4.4 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 1.5 | 11.7 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 1.2 | 3.7 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 1.2 | 6.0 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 1.2 | 17.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 1.1 | 7.9 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) oxidative stress-induced premature senescence(GO:0090403) |
| 1.1 | 20.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) cellular response to low-density lipoprotein particle stimulus(GO:0071404) |
| 1.1 | 6.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 1.0 | 11.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 1.0 | 3.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.9 | 3.8 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.9 | 3.8 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.9 | 8.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.9 | 7.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) nucleosome positioning(GO:0016584) |
| 0.9 | 2.6 | GO:1990869 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.8 | 4.8 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.8 | 15.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.8 | 5.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.8 | 4.6 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.8 | 0.8 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.8 | 10.6 | GO:0036065 | fucosylation(GO:0036065) |
| 0.7 | 10.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.7 | 12.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.7 | 22.2 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.7 | 12.0 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.7 | 8.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.7 | 24.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.6 | 5.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.6 | 5.5 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.6 | 6.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.6 | 9.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.6 | 14.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.6 | 2.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.6 | 9.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.6 | 24.8 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.6 | 9.4 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.5 | 4.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.5 | 16.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.5 | 6.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.5 | 6.3 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.4 | 3.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.4 | 5.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.4 | 6.0 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.4 | 2.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.4 | 4.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.4 | 4.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.4 | 18.8 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.4 | 34.1 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.4 | 6.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.4 | 4.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.4 | 9.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.4 | 1.1 | GO:0090648 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) response to environmental enrichment(GO:0090648) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.4 | 1.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.3 | 1.2 | GO:2000293 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.3 | 2.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 4.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.3 | 6.1 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.3 | 2.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 2.2 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.2 | 8.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 13.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 0.4 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.2 | 1.7 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.2 | 10.9 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.2 | 1.6 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.2 | 2.9 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.2 | 4.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 11.2 | GO:0008542 | visual learning(GO:0008542) |
| 0.2 | 1.8 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 1.0 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 4.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 11.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 3.7 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 11.0 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.1 | 16.2 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 1.5 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 2.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.3 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 9.0 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 4.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.3 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 1.8 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 4.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 8.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 1.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 2.0 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 1.8 | GO:0032680 | regulation of tumor necrosis factor production(GO:0032680) |
| 0.0 | 9.6 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 5.3 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 3.1 | GO:2001234 | negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 | 0.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.0 | GO:0070110 | interleukin-6 receptor complex(GO:0005896) ciliary neurotrophic factor receptor complex(GO:0070110) |
| 2.4 | 12.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 2.3 | 14.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 1.4 | 5.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.3 | 3.9 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 1.3 | 45.2 | GO:0008305 | integrin complex(GO:0008305) |
| 1.1 | 5.5 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.0 | 9.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 1.0 | 11.0 | GO:0042555 | MCM complex(GO:0042555) |
| 1.0 | 4.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.0 | 8.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.0 | 10.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.9 | 18.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.9 | 6.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.9 | 6.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.8 | 15.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.6 | 4.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.6 | 6.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.6 | 7.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.6 | 4.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.5 | 6.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.5 | 70.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.5 | 27.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.5 | 3.8 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.5 | 45.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.4 | 2.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.4 | 6.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.4 | 7.0 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.4 | 24.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.4 | 5.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 22.0 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.3 | 33.4 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.3 | 24.2 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.3 | 3.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 1.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 4.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 12.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 11.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.2 | 13.5 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.2 | 13.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 3.8 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.2 | 62.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 14.0 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.2 | 18.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 18.2 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 17.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.2 | 1.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.8 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 3.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 24.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 7.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 19.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 30.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 7.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 34.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 1.2 | GO:0070852 | varicosity(GO:0043196) cell body fiber(GO:0070852) |
| 0.1 | 4.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 14.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 9.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 14.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.7 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 7.2 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 3.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 0.5 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 12.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 3.1 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 7.1 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 1.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 9.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 2.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 8.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 5.2 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 5.9 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 5.4 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 6.5 | 19.6 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 5.3 | 21.2 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 3.8 | 11.4 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 3.8 | 11.3 | GO:0002060 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 3.5 | 10.6 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 3.3 | 13.0 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 3.1 | 9.4 | GO:0043398 | HLH domain binding(GO:0043398) |
| 2.4 | 12.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 2.3 | 9.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 2.2 | 15.2 | GO:0035375 | zymogen binding(GO:0035375) |
| 2.0 | 36.8 | GO:0005522 | profilin binding(GO:0005522) |
| 1.9 | 11.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.9 | 9.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 1.8 | 5.5 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.8 | 5.5 | GO:0004040 | amidase activity(GO:0004040) |
| 1.7 | 38.1 | GO:0051861 | glycolipid binding(GO:0051861) |
| 1.6 | 17.3 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 1.3 | 3.9 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 1.3 | 7.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 1.2 | 3.7 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 1.1 | 5.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.1 | 4.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 1.0 | 10.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.0 | 13.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.9 | 3.8 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.9 | 17.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.9 | 14.0 | GO:0005536 | glucose binding(GO:0005536) |
| 0.9 | 52.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.8 | 21.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.7 | 2.9 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.7 | 17.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.6 | 18.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.6 | 3.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 6.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.5 | 4.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.5 | 6.1 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.5 | 5.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.4 | 61.3 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.4 | 6.3 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.4 | 7.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.4 | 9.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 6.9 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.3 | 21.2 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.3 | 1.2 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.3 | 8.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.3 | 24.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.3 | 13.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.3 | 6.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 1.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 8.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 4.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 1.1 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) MRF binding(GO:0043426) |
| 0.2 | 21.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 2.6 | GO:0035925 | miRNA binding(GO:0035198) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 14.5 | GO:0051287 | NAD binding(GO:0051287) |
| 0.2 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 7.1 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.2 | 4.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 5.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 44.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.2 | 17.7 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.1 | 9.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 11.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 4.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 4.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 31.6 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.3 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 0.1 | 4.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.5 | GO:0004935 | adrenergic receptor activity(GO:0004935) |
| 0.1 | 3.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 3.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 9.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.1 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 1.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 37.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0047631 | adenosine-diphosphatase activity(GO:0043262) ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 4.8 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 2.5 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 6.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.4 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 11.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 5.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 3.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 35.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 2.6 | 47.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.6 | 45.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 1.0 | 23.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.7 | 9.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.7 | 14.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.6 | 32.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.5 | 6.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 19.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 9.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 7.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.3 | 18.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 5.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 6.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 4.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 3.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 14.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 1.7 | 19.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 1.7 | 78.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 1.7 | 17.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.0 | 19.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.9 | 11.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.8 | 14.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.8 | 24.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.7 | 9.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.6 | 19.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.6 | 11.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.5 | 17.5 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.5 | 11.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 4.4 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.4 | 4.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 11.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.4 | 8.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.3 | 9.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 9.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 4.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.3 | 3.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.3 | 3.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 10.4 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.2 | 6.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.2 | 5.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 7.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 7.6 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 1.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 2.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 14.0 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.1 | 6.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 2.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |