Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
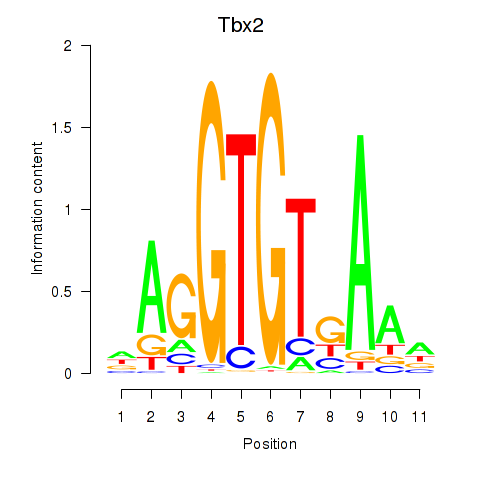
Results for Tbx2
Z-value: 0.75
Transcription factors associated with Tbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx2
|
ENSRNOG00000003517 | T-box 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx2 | rn6_v1_chr10_+_73279119_73279119 | 0.01 | 8.2e-01 | Click! |
Activity profile of Tbx2 motif
Sorted Z-values of Tbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_164808323 | 51.76 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr2_-_45077219 | 47.51 |
ENSRNOT00000014319
|
Gzmk
|
granzyme K |
| chr8_-_107952530 | 24.52 |
ENSRNOT00000052043
|
Cldn18
|
claudin 18 |
| chr1_-_197821936 | 24.03 |
ENSRNOT00000055027
|
Cd19
|
CD19 molecule |
| chr8_+_58431407 | 19.08 |
ENSRNOT00000011974
|
Sln
|
sarcolipin |
| chr1_+_196996581 | 18.67 |
ENSRNOT00000021690
|
Il21r
|
interleukin 21 receptor |
| chrX_+_134979646 | 18.28 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chrX_+_128897181 | 18.16 |
ENSRNOT00000008444
|
Sh2d1a
|
SH2 domain containing 1A |
| chr1_+_87938042 | 16.26 |
ENSRNOT00000027837
|
Map4k1
|
mitogen activated protein kinase kinase kinase kinase 1 |
| chr14_-_14390699 | 16.00 |
ENSRNOT00000046639
|
Anxa3
|
annexin A3 |
| chr1_-_20155960 | 15.14 |
ENSRNOT00000061389
|
Samd3
|
sterile alpha motif domain containing 3 |
| chr16_-_49574314 | 14.63 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr13_-_85443976 | 11.22 |
ENSRNOT00000005213
|
Uck2
|
uridine-cytidine kinase 2 |
| chrX_+_70563570 | 11.05 |
ENSRNOT00000003772
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr7_-_12635943 | 10.97 |
ENSRNOT00000015029
|
Cfd
|
complement factor D |
| chr15_+_28295368 | 10.19 |
ENSRNOT00000013786
|
Slc39a2
|
solute carrier family 39 member 2 |
| chr5_+_24434872 | 9.57 |
ENSRNOT00000010696
|
Ccne2
|
cyclin E2 |
| chr20_-_4677034 | 9.56 |
ENSRNOT00000075244
|
Rn60_20_0047.3
|
|
| chrX_-_62690806 | 8.99 |
ENSRNOT00000018147
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr9_+_98505259 | 8.18 |
ENSRNOT00000051810
|
Erfe
|
erythroferrone |
| chr3_+_170994038 | 8.06 |
ENSRNOT00000081823
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chr13_-_47154292 | 8.04 |
ENSRNOT00000005284
|
Cd55
|
CD55 molecule, decay accelerating factor for complement |
| chr9_-_78693028 | 7.87 |
ENSRNOT00000036359
|
Abca12
|
ATP binding cassette subfamily A member 12 |
| chr1_-_124803363 | 7.87 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr10_+_49541051 | 7.85 |
ENSRNOT00000041606
|
Pmp22
|
peripheral myelin protein 22 |
| chr13_-_85443727 | 7.69 |
ENSRNOT00000090668
ENSRNOT00000076125 |
Uck2
|
uridine-cytidine kinase 2 |
| chr4_-_155421998 | 7.42 |
ENSRNOT00000041552
|
Gdf3
|
growth differentiation factor 3 |
| chr6_-_142880382 | 6.86 |
ENSRNOT00000074706
ENSRNOT00000072861 |
AABR07065821.1
|
|
| chr12_-_3924415 | 6.69 |
ENSRNOT00000067752
|
AABR07035074.1
|
|
| chr3_+_153580861 | 6.46 |
ENSRNOT00000080516
ENSRNOT00000012739 |
Src
|
SRC proto-oncogene, non-receptor tyrosine kinase |
| chr13_+_82429063 | 6.38 |
ENSRNOT00000076879
|
Selp
|
selectin P |
| chr2_-_251532312 | 6.36 |
ENSRNOT00000019501
|
Cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr20_-_4677506 | 6.30 |
ENSRNOT00000061297
|
RT1-CE6
|
RT1 class I, locus CE6 |
| chr6_-_143206772 | 6.18 |
ENSRNOT00000073713
|
AABR07065839.1
|
|
| chr3_-_66335869 | 6.13 |
ENSRNOT00000079781
ENSRNOT00000043238 ENSRNOT00000073412 ENSRNOT00000057878 ENSRNOT00000079212 |
Cerkl
|
ceramide kinase-like |
| chr19_+_39063998 | 5.94 |
ENSRNOT00000081116
|
Has3
|
hyaluronan synthase 3 |
| chr19_+_33932532 | 5.86 |
ENSRNOT00000078395
ENSRNOT00000091985 ENSRNOT00000078701 ENSRNOT00000080317 |
Ednra
|
endothelin receptor type A |
| chr12_+_41385241 | 5.77 |
ENSRNOT00000074974
|
Dtx1
|
deltex E3 ubiquitin ligase 1 |
| chr17_-_15467320 | 5.76 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chr8_-_45215974 | 5.69 |
ENSRNOT00000010792
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr4_+_49369296 | 5.60 |
ENSRNOT00000007822
|
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chrX_+_104734082 | 5.49 |
ENSRNOT00000005020
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr17_-_89163113 | 5.41 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr6_+_109300433 | 5.39 |
ENSRNOT00000010712
|
Fos
|
FBJ osteosarcoma oncogene |
| chr15_+_30735921 | 4.95 |
ENSRNOT00000078887
|
AABR07017763.2
|
|
| chr19_-_37907714 | 4.68 |
ENSRNOT00000026361
|
Ctrl
|
chymotrypsin-like |
| chr10_+_63778755 | 4.57 |
ENSRNOT00000090481
|
Inpp5k
|
inositol polyphosphate-5-phosphatase K |
| chr1_-_214423881 | 4.57 |
ENSRNOT00000025290
|
Pidd1
|
p53-induced death domain protein 1 |
| chr20_-_3728844 | 4.24 |
ENSRNOT00000074958
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 |
| chr3_-_164388492 | 4.16 |
ENSRNOT00000090493
|
Tmem189
|
transmembrane protein 189 |
| chr1_+_22332090 | 4.12 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr1_-_141533908 | 4.08 |
ENSRNOT00000020117
|
Mesp1
|
mesoderm posterior bHLH transcription factor 1 |
| chr1_-_172567846 | 3.89 |
ENSRNOT00000012974
|
Olr259
|
olfactory receptor 259 |
| chr7_-_62972084 | 3.81 |
ENSRNOT00000064251
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chr10_-_56511583 | 3.81 |
ENSRNOT00000021402
|
LOC497940
|
similar to RIKEN cDNA 2810408A11 |
| chr8_-_75995371 | 3.81 |
ENSRNOT00000014014
|
Foxb1
|
forkhead box B1 |
| chr5_-_171415354 | 3.62 |
ENSRNOT00000055398
|
Tp73
|
tumor protein p73 |
| chr1_+_141550633 | 3.52 |
ENSRNOT00000020047
|
Mesp2
|
mesoderm posterior bHLH transcription factor 2 |
| chr5_-_57845509 | 3.38 |
ENSRNOT00000035541
|
Kif24
|
kinesin family member 24 |
| chr6_-_11860001 | 3.30 |
ENSRNOT00000052280
|
H2afz
|
H2A histone family, member Z |
| chr14_-_21709084 | 3.23 |
ENSRNOT00000087477
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chr1_+_85003280 | 3.02 |
ENSRNOT00000057122
|
Fcgbp
|
Fc fragment of IgG binding protein |
| chr19_+_12097632 | 2.94 |
ENSRNOT00000049452
|
Ces5a
|
carboxylesterase 5A |
| chr5_-_140834950 | 2.48 |
ENSRNOT00000043799
ENSRNOT00000089980 |
Ppie
|
peptidylprolyl isomerase E |
| chr1_+_187149453 | 2.48 |
ENSRNOT00000082738
|
Xylt1
|
xylosyltransferase 1 |
| chr6_-_50923510 | 2.42 |
ENSRNOT00000010631
|
Bcap29
|
B-cell receptor-associated protein 29 |
| chr6_-_103605256 | 2.41 |
ENSRNOT00000006005
|
Dcaf5
|
DDB1 and CUL4 associated factor 5 |
| chr4_-_55527092 | 2.40 |
ENSRNOT00000087407
ENSRNOT00000010223 |
Zfp800
|
zinc finger protein 800 |
| chrX_+_33599671 | 2.38 |
ENSRNOT00000006843
|
Txlng
|
taxilin gamma |
| chr20_+_6356423 | 2.38 |
ENSRNOT00000000628
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr18_+_72571327 | 2.37 |
ENSRNOT00000089468
|
Smad2
|
SMAD family member 2 |
| chr1_+_168378637 | 2.27 |
ENSRNOT00000028936
|
Olr86
|
olfactory receptor 86 |
| chr7_-_11406771 | 2.23 |
ENSRNOT00000047450
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr17_+_72240802 | 2.18 |
ENSRNOT00000025993
ENSRNOT00000090453 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr5_+_50381244 | 2.00 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr1_+_22364551 | 1.99 |
ENSRNOT00000043157
|
LOC108348200
|
trace amine-associated receptor 8a |
| chr19_-_31867850 | 1.97 |
ENSRNOT00000024689
|
Anapc10
|
anaphase promoting complex subunit 10 |
| chr2_+_140471690 | 1.89 |
ENSRNOT00000017379
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr17_-_14828329 | 1.87 |
ENSRNOT00000022830
|
Barx1
|
BARX homeobox 1 |
| chr8_+_41285450 | 1.77 |
ENSRNOT00000076562
|
Olr1222
|
olfactory receptor 1222 |
| chr5_+_159671136 | 1.64 |
ENSRNOT00000084737
|
Fbxo42
|
F-box protein 42 |
| chr4_-_145678066 | 1.63 |
ENSRNOT00000014103
|
Ghrl
|
ghrelin and obestatin prepropeptide |
| chr16_+_61130755 | 1.63 |
ENSRNOT00000042609
|
AABR07026048.1
|
|
| chr5_-_171710312 | 1.57 |
ENSRNOT00000076044
ENSRNOT00000071280 ENSRNOT00000072059 ENSRNOT00000077015 |
Prdm16
|
PR/SET domain 16 |
| chr2_+_211880262 | 1.56 |
ENSRNOT00000038315
|
Slc25a54
|
solute carrier family 25, member 54 |
| chr18_+_69549937 | 1.48 |
ENSRNOT00000041227
|
Mex3c
|
mex-3 RNA binding family member C |
| chr3_-_94419048 | 1.47 |
ENSRNOT00000015775
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr4_+_1584083 | 1.45 |
ENSRNOT00000087099
|
Olr1244
|
olfactory receptor 1244 |
| chr5_+_113592919 | 1.44 |
ENSRNOT00000011336
|
Ift74
|
intraflagellar transport 74 |
| chr18_+_16616937 | 1.41 |
ENSRNOT00000093641
|
Mocos
|
molybdenum cofactor sulfurase |
| chr1_-_142500367 | 1.40 |
ENSRNOT00000015948
ENSRNOT00000089591 |
Crtc3
|
CREB regulated transcription coactivator 3 |
| chr2_+_198755262 | 1.21 |
ENSRNOT00000028807
|
Rbm8a
|
RNA binding motif protein 8A |
| chr7_+_143679617 | 1.20 |
ENSRNOT00000014033
|
Eif4b
|
eukaryotic translation initiation factor 4B |
| chr1_+_100224401 | 1.19 |
ENSRNOT00000025846
|
Fv1
|
Friend virus susceptibility 1 |
| chr4_+_71675383 | 1.17 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr8_+_118744229 | 1.05 |
ENSRNOT00000028364
|
Kif9
|
kinesin family member 9 |
| chr10_-_88645364 | 0.98 |
ENSRNOT00000030344
|
Rab5c
|
RAB5C, member RAS oncogene family |
| chr4_+_1658278 | 0.96 |
ENSRNOT00000073845
|
Olr1250
|
olfactory receptor 1250 |
| chr7_-_126382449 | 0.84 |
ENSRNOT00000085540
|
7530416G11Rik
|
RIKEN cDNA 7530416G11 gene |
| chr8_+_41936959 | 0.79 |
ENSRNOT00000042601
|
Olr1222
|
olfactory receptor 1222 |
| chr7_-_120839577 | 0.74 |
ENSRNOT00000018526
|
Dmc1
|
DNA meiotic recombinase 1 |
| chr1_-_22404002 | 0.49 |
ENSRNOT00000044098
|
Taar8a
|
trace amine-associated receptor 8a |
| chr19_-_25288335 | 0.44 |
ENSRNOT00000050214
|
Nanos3
|
nanos C2HC-type zinc finger 3 |
| chr1_-_172616633 | 0.43 |
ENSRNOT00000012919
|
Olr262
|
olfactory receptor 262 |
| chr3_-_149563476 | 0.37 |
ENSRNOT00000071869
|
AABR07054352.2
|
|
| chr9_+_21045457 | 0.34 |
ENSRNOT00000031759
|
Opn5
|
opsin 5 |
| chr11_-_61530567 | 0.33 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr9_+_70310932 | 0.29 |
ENSRNOT00000088268
|
LOC103690541
|
uncharacterized LOC103690541 |
| chr7_-_141624972 | 0.23 |
ENSRNOT00000078785
|
AABR07058884.1
|
|
| chr18_+_25613831 | 0.22 |
ENSRNOT00000091040
|
Tslp
|
thymic stromal lymphopoietin |
| chr7_-_122926336 | 0.20 |
ENSRNOT00000000205
|
Chadl
|
chondroadherin-like |
| chr1_+_168623150 | 0.16 |
ENSRNOT00000021282
|
Olr107
|
olfactory receptor 107 |
| chr11_+_75905443 | 0.14 |
ENSRNOT00000002650
|
Fgf12
|
fibroblast growth factor 12 |
| chr1_-_169386939 | 0.04 |
ENSRNOT00000023007
|
Olr150
|
olfactory receptor 150 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.3 | 51.8 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 8.2 | 24.5 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 4.7 | 18.9 | GO:0044211 | CTP salvage(GO:0044211) |
| 3.0 | 42.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 2.7 | 19.1 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 2.2 | 9.0 | GO:0006272 | leading strand elongation(GO:0006272) |
| 2.0 | 8.0 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 1.6 | 8.1 | GO:1990918 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 1.6 | 7.9 | GO:0035627 | ceramide transport(GO:0035627) |
| 1.5 | 5.9 | GO:0036118 | hyaluranon cable assembly(GO:0036118) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.4 | 4.1 | GO:0060974 | cell migration involved in heart formation(GO:0060974) regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 1.3 | 18.3 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 1.3 | 6.5 | GO:0051902 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) negative regulation of mitochondrial depolarization(GO:0051902) cellular response to progesterone stimulus(GO:0071393) |
| 1.3 | 3.8 | GO:0030091 | protein repair(GO:0030091) |
| 1.2 | 3.6 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 1.2 | 5.9 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 1.1 | 11.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.1 | 6.4 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 1.0 | 11.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 1.0 | 16.0 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 1.0 | 16.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 1.0 | 3.8 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.9 | 4.6 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.8 | 7.4 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.8 | 5.6 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.8 | 6.4 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.7 | 3.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.6 | 2.4 | GO:1900224 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.6 | 6.9 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.6 | 5.7 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.6 | 7.9 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.5 | 5.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.5 | 7.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 2.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.4 | 8.2 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.4 | 1.6 | GO:0051464 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) positive regulation of cortisol secretion(GO:0051464) |
| 0.4 | 2.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.4 | 1.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.4 | 10.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.3 | 1.9 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 5.4 | GO:0009629 | response to gravity(GO:0009629) |
| 0.3 | 1.5 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.2 | 2.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 1.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 1.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 | 2.0 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.2 | 10.3 | GO:0007129 | synapsis(GO:0007129) |
| 0.2 | 1.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 1.6 | GO:0015866 | ADP transport(GO:0015866) |
| 0.2 | 2.0 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 5.8 | GO:0045581 | negative regulation of T cell differentiation(GO:0045581) |
| 0.1 | 1.0 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 1.4 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 1.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 6.1 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.1 | 5.8 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 2.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.5 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 3.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 14.6 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 3.2 | GO:0051931 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 3.3 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 18.7 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 1.1 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.3 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 1.7 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 2.4 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.2 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 1.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:1905150 | regulation of neuronal action potential(GO:0098908) regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 34.6 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 3.4 | GO:0007018 | microtubule-based movement(GO:0007018) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 1.8 | 51.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 1.6 | 6.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 1.3 | 7.9 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 1.1 | 9.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.1 | 5.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.9 | 16.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.8 | 2.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.6 | 2.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.5 | 19.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 3.3 | GO:0001740 | Barr body(GO:0001740) |
| 0.3 | 2.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 7.8 | GO:0043218 | compact myelin(GO:0043218) |
| 0.3 | 2.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.3 | 3.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 2.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 9.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 7.5 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 22.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 6.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 11.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 5.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 14.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 2.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 32.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 2.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 4.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 18.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 3.9 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 2.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 13.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.6 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 18.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 3.2 | 16.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 2.7 | 16.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 2.1 | 51.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 2.0 | 8.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.8 | 11.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.6 | 6.4 | GO:0042806 | fucose binding(GO:0042806) |
| 1.5 | 4.6 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 1.3 | 7.9 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 1.3 | 9.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 1.2 | 6.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 1.2 | 5.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.2 | 5.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 1.0 | 3.8 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.8 | 2.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.8 | 2.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.7 | 18.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.6 | 2.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.5 | 4.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 8.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.4 | 1.6 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.4 | 1.2 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.4 | 1.4 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 0.4 | 10.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 14.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.3 | 9.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.3 | 63.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 6.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 3.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 5.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 18.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.2 | 1.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 2.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 2.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 7.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 3.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 2.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 5.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 24.0 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 3.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 2.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 3.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 12.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 5.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 2.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 4.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.6 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 15.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 23.5 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 2.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.9 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.0 | GO:0019003 | GDP binding(GO:0019003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 16.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.7 | 9.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.7 | 57.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.6 | 24.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.5 | 6.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.3 | 2.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 15.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 2.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 4.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 7.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.2 | 6.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 6.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 18.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 16.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 5.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 5.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 3.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 3.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 3.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 3.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 51.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.8 | 11.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.7 | 24.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.7 | 6.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.7 | 18.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.6 | 24.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.6 | 8.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.5 | 10.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.5 | 9.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.4 | 7.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 5.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 9.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 5.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.3 | 5.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 12.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 2.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 4.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 2.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 6.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 3.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 3.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 5.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 7.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 2.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |