Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Tbx1_Eomes
Z-value: 0.67
Transcription factors associated with Tbx1_Eomes
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
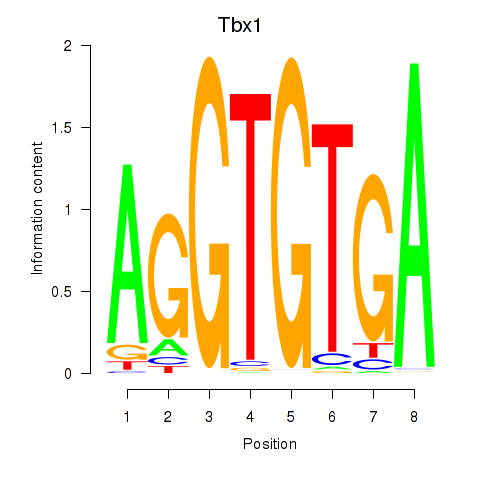
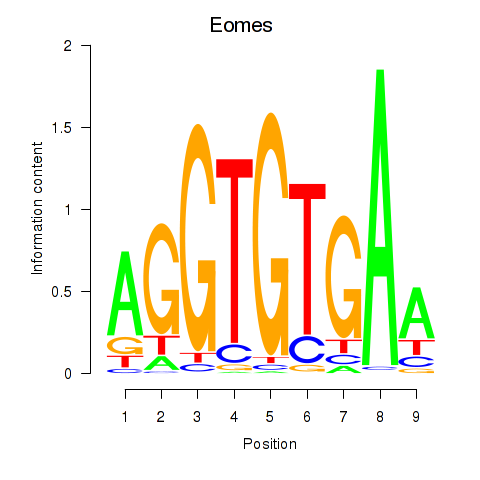
Tbx1
|
ENSRNOG00000001892 | T-box 1 |
|
Eomes
|
ENSRNOG00000042140 | eomesodermin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Eomes | rn6_v1_chr8_+_127144903_127144903 | 0.23 | 4.6e-05 | Click! |
| Tbx1 | rn6_v1_chr11_+_86552022_86552022 | 0.02 | 6.6e-01 | Click! |
Activity profile of Tbx1_Eomes motif
Sorted Z-values of Tbx1_Eomes motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_52350624 | 53.76 |
ENSRNOT00000060476
|
Tmem229a
|
transmembrane protein 229A |
| chr5_+_143500441 | 44.27 |
ENSRNOT00000045513
|
Grik3
|
glutamate ionotropic receptor kainate type subunit 3 |
| chr5_+_164808323 | 42.18 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chrX_-_142248369 | 41.51 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr5_-_12563429 | 29.76 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr17_-_80320681 | 27.61 |
ENSRNOT00000023637
|
C1ql3
|
complement C1q like 3 |
| chr2_+_263212051 | 20.88 |
ENSRNOT00000089396
|
Negr1
|
neuronal growth regulator 1 |
| chr11_-_4397361 | 18.68 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr5_+_136683592 | 18.07 |
ENSRNOT00000085527
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr2_+_74360622 | 18.07 |
ENSRNOT00000014013
|
Cdh18
|
cadherin 18 |
| chr4_-_148845267 | 17.52 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr16_+_46731403 | 16.54 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr14_+_12218553 | 15.56 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr1_+_187149453 | 14.27 |
ENSRNOT00000082738
|
Xylt1
|
xylosyltransferase 1 |
| chr18_+_46148849 | 13.78 |
ENSRNOT00000026724
|
Prr16
|
proline rich 16 |
| chr13_-_25262469 | 13.50 |
ENSRNOT00000019921
|
Rnf152
|
ring finger protein 152 |
| chr10_-_88667345 | 12.80 |
ENSRNOT00000025518
|
Kcnh4
|
potassium voltage-gated channel subfamily H member 4 |
| chr1_-_165680176 | 12.43 |
ENSRNOT00000025245
ENSRNOT00000082697 |
Plekhb1
|
pleckstrin homology domain containing B1 |
| chr3_+_161433410 | 12.38 |
ENSRNOT00000024657
|
Slc12a5
|
solute carrier family 12 member 5 |
| chr10_+_86657285 | 12.02 |
ENSRNOT00000087346
|
Thra
|
thyroid hormone receptor alpha |
| chr3_+_131351587 | 11.92 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr10_+_85608544 | 11.84 |
ENSRNOT00000092122
|
Mllt6
|
MLLT6, PHD finger domain containing |
| chr9_-_63637677 | 11.57 |
ENSRNOT00000049259
|
Satb2
|
SATB homeobox 2 |
| chr8_+_28352772 | 9.83 |
ENSRNOT00000012391
|
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr6_-_124429701 | 9.75 |
ENSRNOT00000037902
ENSRNOT00000080950 |
Ttc7b
|
tetratricopeptide repeat domain 7B |
| chr1_-_226353611 | 9.57 |
ENSRNOT00000037624
|
Dagla
|
diacylglycerol lipase, alpha |
| chr13_+_42220251 | 9.10 |
ENSRNOT00000078185
|
AABR07020815.1
|
|
| chr3_+_100768637 | 8.78 |
ENSRNOT00000083542
|
Bdnf
|
brain-derived neurotrophic factor |
| chr1_+_234363994 | 8.70 |
ENSRNOT00000018137
|
Rorb
|
RAR-related orphan receptor B |
| chr1_+_88542035 | 8.56 |
ENSRNOT00000032973
|
Zfp420
|
zinc finger protein 420 |
| chr4_-_180234804 | 7.99 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr18_+_53609214 | 7.82 |
ENSRNOT00000077443
|
Slc27a6
|
solute carrier family 27 member 6 |
| chr3_+_160231914 | 7.39 |
ENSRNOT00000014411
|
Kcnk15
|
potassium two pore domain channel subfamily K member 15 |
| chr4_+_58053041 | 7.28 |
ENSRNOT00000072698
|
Mest
|
mesoderm specific transcript |
| chr1_+_79982639 | 6.80 |
ENSRNOT00000071916
|
Dmwd
|
dystrophia myotonica, WD repeat containing |
| chr4_+_146455332 | 6.70 |
ENSRNOT00000009775
|
Hrh1
|
histamine receptor H 1 |
| chr10_-_71383378 | 6.46 |
ENSRNOT00000077025
|
Dusp14
|
dual specificity phosphatase 14 |
| chr2_-_62634785 | 6.25 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr8_-_45215974 | 6.24 |
ENSRNOT00000010792
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr12_-_46920952 | 4.91 |
ENSRNOT00000001532
|
Msi1
|
musashi RNA-binding protein 1 |
| chr15_+_8739589 | 4.62 |
ENSRNOT00000071751
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr18_-_28871849 | 4.48 |
ENSRNOT00000025902
|
Nrg2
|
neuregulin 2 |
| chr2_+_140836006 | 4.47 |
ENSRNOT00000074141
|
Dusp14l1
|
dual specificity phosphatase 14-like 1 |
| chr10_-_74001895 | 4.08 |
ENSRNOT00000005658
|
Vmp1
|
vacuole membrane protein 1 |
| chr18_-_40134504 | 3.86 |
ENSRNOT00000022294
|
Trim36
|
tripartite motif-containing 36 |
| chr1_+_224824799 | 3.86 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chr13_-_55878094 | 3.68 |
ENSRNOT00000014218
|
Lhx9
|
LIM homeobox 9 |
| chr10_+_110445797 | 3.54 |
ENSRNOT00000054920
|
Narf
|
nuclear prelamin A recognition factor |
| chr6_+_101532518 | 3.41 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr1_+_89017479 | 3.40 |
ENSRNOT00000038154
|
U2af1l4
|
U2 small nuclear RNA auxiliary factor 1-like 4 |
| chr17_-_14091766 | 3.32 |
ENSRNOT00000075373
|
AABR07027088.1
|
|
| chr10_-_91186054 | 3.28 |
ENSRNOT00000004140
|
Plcd3
|
phospholipase C, delta 3 |
| chr8_-_86286991 | 3.19 |
ENSRNOT00000046322
|
LOC367117
|
similar to RIKEN cDNA 2900055D03 |
| chr10_-_88645364 | 3.14 |
ENSRNOT00000030344
|
Rab5c
|
RAB5C, member RAS oncogene family |
| chr1_-_227047290 | 3.12 |
ENSRNOT00000037226
|
Ms4a15
|
membrane spanning 4-domains A15 |
| chrX_+_16946207 | 3.08 |
ENSRNOT00000003981
|
RGD1563606
|
similar to cysteine-rich protein 2 |
| chr2_+_207262934 | 3.07 |
ENSRNOT00000016833
|
Ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr7_-_120839577 | 2.95 |
ENSRNOT00000018526
|
Dmc1
|
DNA meiotic recombinase 1 |
| chr4_+_71675383 | 2.64 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr1_-_172322795 | 2.58 |
ENSRNOT00000075318
|
RGD1562400
|
similar to olfactory receptor MOR204-14 |
| chr1_+_172822544 | 2.51 |
ENSRNOT00000090018
|
AABR07004960.1
|
|
| chr12_+_52359310 | 2.46 |
ENSRNOT00000071857
ENSRNOT00000065893 |
Fbrsl1
|
fibrosin-like 1 |
| chr6_-_44363915 | 2.42 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr10_-_83104173 | 2.06 |
ENSRNOT00000031252
|
Kat7
|
lysine acetyltransferase 7 |
| chr3_-_43119159 | 2.05 |
ENSRNOT00000041394
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr8_-_36410612 | 1.99 |
ENSRNOT00000091308
|
Foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chrX_+_70563570 | 1.90 |
ENSRNOT00000003772
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr1_+_150225373 | 1.81 |
ENSRNOT00000051266
|
Olr30
|
olfactory receptor 30 |
| chr3_+_2490518 | 1.79 |
ENSRNOT00000015099
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr8_-_43443531 | 1.67 |
ENSRNOT00000007901
|
LOC103693060
|
olfactory receptor 148-like |
| chr5_+_58393603 | 1.62 |
ENSRNOT00000080082
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr5_-_819326 | 1.59 |
ENSRNOT00000023899
|
Pi15
|
peptidase inhibitor 15 |
| chr13_-_90467235 | 1.49 |
ENSRNOT00000081534
ENSRNOT00000008150 |
Ncstn
|
nicastrin |
| chr1_+_85485289 | 1.43 |
ENSRNOT00000026182
|
Dll3
|
delta like canonical Notch ligand 3 |
| chrX_+_128897181 | 1.42 |
ENSRNOT00000008444
|
Sh2d1a
|
SH2 domain containing 1A |
| chr7_+_43601513 | 1.29 |
ENSRNOT00000030321
|
AABR07056845.1
|
|
| chr7_-_27240528 | 1.29 |
ENSRNOT00000029435
ENSRNOT00000079731 |
Hsp90b1
|
heat shock protein 90 beta family member 1 |
| chr11_+_43143882 | 1.19 |
ENSRNOT00000041445
|
Olr1528
|
olfactory receptor 1528 |
| chr10_-_86393141 | 1.05 |
ENSRNOT00000009485
|
Mien1
|
migration and invasion enhancer 1 |
| chr1_+_7252349 | 0.86 |
ENSRNOT00000030329
|
Plagl1
|
PLAG1 like zinc finger 1 |
| chrM_+_7758 | 0.85 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr19_+_39126589 | 0.85 |
ENSRNOT00000027561
|
Sntb2
|
syntrophin, beta 2 |
| chrM_+_7919 | 0.63 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr9_-_15700235 | 0.57 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr5_+_156587093 | 0.57 |
ENSRNOT00000019696
|
Hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr1_+_172051215 | 0.55 |
ENSRNOT00000048370
|
Olr235
|
olfactory receptor 235 |
| chr4_+_57034675 | 0.51 |
ENSRNOT00000080223
|
Smo
|
smoothened, frizzled class receptor |
| chr5_+_63781801 | 0.45 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr1_-_124803363 | 0.45 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr8_+_41936959 | 0.44 |
ENSRNOT00000042601
|
Olr1222
|
olfactory receptor 1222 |
| chr6_-_50923510 | 0.37 |
ENSRNOT00000010631
|
Bcap29
|
B-cell receptor-associated protein 29 |
| chr1_+_282557426 | 0.32 |
ENSRNOT00000088966
|
AABR07007134.1
|
|
| chr11_-_45616429 | 0.22 |
ENSRNOT00000046587
|
Olr1533
|
olfactory receptor 1533 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx1_Eomes
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.1 | 42.2 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 5.9 | 41.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 5.2 | 15.6 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 4.0 | 44.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 4.0 | 12.0 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 3.9 | 11.8 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 3.2 | 9.6 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 2.2 | 6.7 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 1.8 | 12.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.6 | 18.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 1.5 | 16.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 1.4 | 12.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 1.2 | 3.7 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 1.2 | 14.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 1.1 | 8.8 | GO:0061193 | taste bud development(GO:0061193) |
| 1.1 | 11.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.9 | 3.4 | GO:0097112 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.8 | 2.4 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.8 | 3.9 | GO:0031427 | response to methotrexate(GO:0031427) |
| 0.7 | 8.0 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.7 | 8.7 | GO:0046549 | amacrine cell differentiation(GO:0035881) retinal rod cell development(GO:0046548) retinal cone cell development(GO:0046549) |
| 0.7 | 2.1 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.7 | 2.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.6 | 6.2 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.6 | 9.8 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.6 | 13.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.4 | 3.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.4 | 3.0 | GO:0042148 | strand invasion(GO:0042148) |
| 0.4 | 1.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.3 | 13.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.3 | 36.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 9.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.2 | 4.6 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 | 2.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 29.4 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.2 | 0.6 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.2 | 4.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 1.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 7.3 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.1 | 17.8 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 1.3 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.5 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 3.3 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 1.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 2.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 3.9 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 7.8 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.1 | 0.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 11.9 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 4.1 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 6.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 1.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 4.5 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 1.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 7.4 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 20.8 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 3.3 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 42.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 1.3 | 44.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.7 | 3.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.6 | 41.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.5 | 8.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.5 | 9.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.5 | 18.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.4 | 3.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.4 | 12.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.4 | 13.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 20.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 11.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 11.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 12.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 4.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) autophagosome membrane(GO:0000421) |
| 0.1 | 0.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.5 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 1.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 16.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 4.4 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 6.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 18.1 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 21.4 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 2.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 14.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.0 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 129.5 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 2.1 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 44.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 4.8 | 14.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 3.1 | 15.6 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 1.8 | 12.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.7 | 8.7 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 1.7 | 42.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 1.5 | 16.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 1.4 | 18.1 | GO:0015187 | sodium:amino acid symporter activity(GO:0005283) glycine transmembrane transporter activity(GO:0015187) |
| 1.3 | 6.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 1.1 | 8.0 | GO:0043426 | MRF binding(GO:0043426) |
| 1.1 | 4.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.1 | 8.8 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.0 | 12.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.9 | 41.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.9 | 3.4 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.8 | 7.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.5 | 3.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.5 | 3.9 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.3 | 3.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.3 | 1.9 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.3 | 4.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 4.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 3.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 3.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 2.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 2.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 15.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 0.5 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 52.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 7.4 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.1 | 3.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 3.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 18.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 23.8 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 5.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 42.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 10.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 4.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 9.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 27.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 10.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 8.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 29.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 0.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 44.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.7 | 15.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 1.5 | 42.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 1.3 | 36.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.8 | 18.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 27.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.4 | 3.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 8.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.3 | 9.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 4.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 12.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 7.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 6.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 6.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 12.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 0.8 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 3.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 3.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 3.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 3.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |