Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Tbx15
Z-value: 0.80
Transcription factors associated with Tbx15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx15
|
ENSRNOG00000019565 | T-box 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx15 | rn6_v1_chr2_+_201289357_201289357 | 0.02 | 6.6e-01 | Click! |
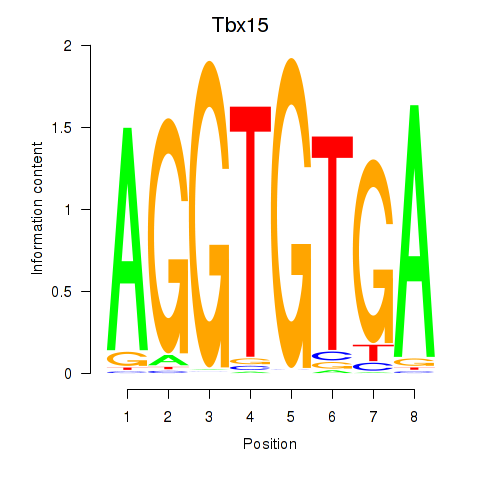
Activity profile of Tbx15 motif
Sorted Z-values of Tbx15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_53609214 | 70.85 |
ENSRNOT00000077443
|
Slc27a6
|
solute carrier family 27 member 6 |
| chr4_-_148845267 | 28.44 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr3_+_61620192 | 21.01 |
ENSRNOT00000065426
|
Hoxd9
|
homeo box D9 |
| chr2_-_23289266 | 19.57 |
ENSRNOT00000061708
|
Bhmt2
|
betaine-homocysteine S-methyltransferase 2 |
| chr1_+_7252349 | 17.31 |
ENSRNOT00000030329
|
Plagl1
|
PLAG1 like zinc finger 1 |
| chr2_+_60920257 | 13.46 |
ENSRNOT00000025170
|
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr11_-_81717521 | 12.45 |
ENSRNOT00000058422
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr6_-_127653124 | 11.32 |
ENSRNOT00000047324
|
Serpina11
|
serpin family A member 11 |
| chr8_-_78233430 | 10.79 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chrX_-_142248369 | 10.77 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr13_-_25262469 | 10.73 |
ENSRNOT00000019921
|
Rnf152
|
ring finger protein 152 |
| chr1_+_224824799 | 10.67 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chr16_+_46731403 | 10.67 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr14_+_12218553 | 10.63 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr1_+_48273611 | 10.51 |
ENSRNOT00000022254
ENSRNOT00000022068 |
Slc22a1
|
solute carrier family 22 member 1 |
| chr1_+_238222521 | 9.49 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chrX_-_139464798 | 9.29 |
ENSRNOT00000003282
|
Gpc4
|
glypican 4 |
| chr18_+_69841053 | 9.16 |
ENSRNOT00000071545
ENSRNOT00000030613 ENSRNOT00000075543 |
Mro
|
maestro |
| chr7_-_29152442 | 8.76 |
ENSRNOT00000079774
|
Mybpc1
|
myosin binding protein C, slow type |
| chr13_+_75105615 | 8.67 |
ENSRNOT00000076653
|
Tp53i3
|
tumor protein p53 inducible protein 3 |
| chr20_-_45126062 | 8.42 |
ENSRNOT00000000720
|
RGD1310495
|
similar to KIAA1919 protein |
| chr2_+_22909569 | 8.28 |
ENSRNOT00000073871
|
Homer1
|
homer scaffolding protein 1 |
| chr12_-_46920952 | 8.09 |
ENSRNOT00000001532
|
Msi1
|
musashi RNA-binding protein 1 |
| chr12_-_41627741 | 8.07 |
ENSRNOT00000001875
|
Sds
|
serine dehydratase |
| chr4_-_82194927 | 7.94 |
ENSRNOT00000072302
|
LOC103692128
|
homeobox protein Hox-A9 |
| chr16_-_62373253 | 7.84 |
ENSRNOT00000034325
|
Tex15
|
testis expressed 15, meiosis and synapsis associated |
| chr1_+_170471238 | 7.73 |
ENSRNOT00000076961
ENSRNOT00000075597 ENSRNOT00000076631 ENSRNOT00000076783 |
Timm10b
Dnhd1
|
translocase of inner mitochondrial membrane 10B dynein heavy chain domain 1 |
| chr8_+_117068582 | 7.64 |
ENSRNOT00000073559
|
Amt
|
aminomethyltransferase |
| chr5_+_33784715 | 7.58 |
ENSRNOT00000035685
|
Slc7a13
|
solute carrier family 7 member 13 |
| chr10_+_86657285 | 7.45 |
ENSRNOT00000087346
|
Thra
|
thyroid hormone receptor alpha |
| chr12_+_19196611 | 6.95 |
ENSRNOT00000001801
|
Azgp1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr1_+_80777014 | 6.93 |
ENSRNOT00000079758
|
Gm19345
|
predicted gene, 19345 |
| chr16_+_18690649 | 6.91 |
ENSRNOT00000015190
|
Mat1a
|
methionine adenosyltransferase 1A |
| chr1_+_30681681 | 6.65 |
ENSRNOT00000015395
|
Rspo3
|
R-spondin 3 |
| chr17_-_10640548 | 6.53 |
ENSRNOT00000064274
|
Simc1
|
SUMO-interacting motifs containing 1 |
| chr17_-_8607494 | 6.48 |
ENSRNOT00000016138
|
Slc25a48
|
solute carrier family 25, member 48 |
| chr13_+_49074644 | 6.21 |
ENSRNOT00000000041
|
Klhdc8a
|
kelch domain containing 8A |
| chr12_-_23661009 | 5.99 |
ENSRNOT00000059451
|
Upk3bl
|
uroplakin 3B-like |
| chr15_-_33527031 | 5.88 |
ENSRNOT00000019979
|
Homez
|
homeobox and leucine zipper encoding |
| chr4_+_58053041 | 5.87 |
ENSRNOT00000072698
|
Mest
|
mesoderm specific transcript |
| chr7_-_62972084 | 5.73 |
ENSRNOT00000064251
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chr9_+_100281339 | 5.70 |
ENSRNOT00000029127
|
Agxt
|
alanine-glyoxylate aminotransferase |
| chr4_+_68849033 | 5.70 |
ENSRNOT00000016912
|
Mgam
|
maltase-glucoamylase |
| chr17_-_10622063 | 5.57 |
ENSRNOT00000022980
|
Simc1
|
SUMO-interacting motifs containing 1 |
| chr11_+_27208564 | 5.51 |
ENSRNOT00000002158
|
Map3k7cl
|
MAP3K7 C-terminal like |
| chr5_-_135561914 | 5.50 |
ENSRNOT00000023178
|
Mmachc
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr2_-_210738378 | 5.42 |
ENSRNOT00000025746
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr2_-_53185926 | 5.41 |
ENSRNOT00000081558
|
Ghr
|
growth hormone receptor |
| chrX_+_15295473 | 5.37 |
ENSRNOT00000009295
|
LOC108348065
|
histone deacetylase 6 |
| chr9_-_63637677 | 5.33 |
ENSRNOT00000049259
|
Satb2
|
SATB homeobox 2 |
| chr5_-_12563429 | 5.32 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr8_-_86286991 | 5.32 |
ENSRNOT00000046322
|
LOC367117
|
similar to RIKEN cDNA 2900055D03 |
| chr2_-_187435323 | 5.31 |
ENSRNOT00000050476
|
Ttc24
|
tetratricopeptide repeat domain 24 |
| chr11_-_32508420 | 5.28 |
ENSRNOT00000002717
|
Kcne1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chrX_-_111102464 | 5.23 |
ENSRNOT00000084176
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr5_-_159962218 | 4.99 |
ENSRNOT00000050729
|
Clcnkb
|
chloride voltage-gated channel Kb |
| chr11_+_74057361 | 4.85 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chr7_+_129595192 | 4.82 |
ENSRNOT00000071151
|
Zdhhc25
|
zinc finger, DHHC-type containing 25 |
| chr13_-_50509916 | 4.79 |
ENSRNOT00000076747
|
Ren
|
renin |
| chr4_+_71675383 | 4.76 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr4_-_70974006 | 4.69 |
ENSRNOT00000020975
|
Trpv5
|
transient receptor potential cation channel, subfamily V, member 5 |
| chr4_-_70973842 | 4.69 |
ENSRNOT00000051687
|
Trpv5
|
transient receptor potential cation channel, subfamily V, member 5 |
| chr19_-_25288335 | 4.30 |
ENSRNOT00000050214
|
Nanos3
|
nanos C2HC-type zinc finger 3 |
| chr2_+_207262934 | 4.28 |
ENSRNOT00000016833
|
Ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr5_-_173680290 | 4.24 |
ENSRNOT00000027564
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr8_+_91464229 | 4.17 |
ENSRNOT00000013249
|
Bckdhb
|
branched chain keto acid dehydrogenase E1 subunit beta |
| chr1_-_81946714 | 4.11 |
ENSRNOT00000027578
|
Grik5
|
glutamate ionotropic receptor kainate type subunit 5 |
| chr4_-_15859132 | 4.06 |
ENSRNOT00000082161
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr2_-_210749991 | 4.05 |
ENSRNOT00000051261
ENSRNOT00000052403 |
Gstm6l
|
glutathione S-transferase, mu 6-like |
| chr10_+_85608544 | 4.01 |
ENSRNOT00000092122
|
Mllt6
|
MLLT6, PHD finger domain containing |
| chr10_-_56511583 | 4.00 |
ENSRNOT00000021402
|
LOC497940
|
similar to RIKEN cDNA 2810408A11 |
| chr11_-_65845418 | 3.97 |
ENSRNOT00000091057
|
Fstl1
|
follistatin-like 1 |
| chr1_-_215373234 | 3.90 |
ENSRNOT00000034299
|
AABR07006055.1
|
|
| chr19_+_25262076 | 3.89 |
ENSRNOT00000030658
|
RGD1306072
|
hypothetical LOC304654 |
| chr12_+_9464026 | 3.89 |
ENSRNOT00000048188
|
Cdx2
|
caudal type homeo box 2 |
| chr16_-_70705128 | 3.86 |
ENSRNOT00000071258
|
Rnf170
|
ring finger protein 170 |
| chr10_+_36554416 | 3.85 |
ENSRNOT00000049730
|
Olr1411
|
olfactory receptor 1411 |
| chr10_-_18574909 | 3.80 |
ENSRNOT00000083090
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr2_+_127845034 | 3.78 |
ENSRNOT00000044804
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr1_-_215350161 | 3.77 |
ENSRNOT00000081144
ENSRNOT00000064050 |
AABR07006049.1
|
|
| chr4_-_157439507 | 3.75 |
ENSRNOT00000081802
|
Ptms
|
parathymosin |
| chr1_-_102780381 | 3.72 |
ENSRNOT00000080132
|
Saa4
|
serum amyloid A4 |
| chr4_+_147037179 | 3.66 |
ENSRNOT00000011292
|
Syn2
|
synapsin II |
| chr1_+_88324339 | 3.66 |
ENSRNOT00000072762
|
LOC102547059
|
zinc finger protein 420-like |
| chrX_+_135348436 | 3.62 |
ENSRNOT00000008868
|
Rab33a
|
RAB33A, member RAS oncogene family |
| chr14_-_80973456 | 3.62 |
ENSRNOT00000013257
|
Hgfac
|
HGF activator |
| chr1_+_87903783 | 3.61 |
ENSRNOT00000074299
|
Zfp84
|
zinc finger protein 84 |
| chr10_-_74119009 | 3.60 |
ENSRNOT00000085712
ENSRNOT00000006926 |
Dhx40
|
DEAH-box helicase 40 |
| chr5_+_127925726 | 3.52 |
ENSRNOT00000014401
|
Coa7
|
cytochrome c oxidase assembly factor 7 |
| chr3_-_113423149 | 3.51 |
ENSRNOT00000021008
|
Serinc4
|
serine incorporator 4 |
| chr4_+_82370997 | 3.50 |
ENSRNOT00000072934
|
NEWGENE_1565788
|
even-skipped homeobox 1 |
| chr20_-_4366957 | 3.48 |
ENSRNOT00000088244
|
Rnf5
|
ring finger protein 5 |
| chr6_-_25616995 | 3.45 |
ENSRNOT00000077894
|
Fosl2
|
FOS like 2, AP-1 transcription factor subunit |
| chr10_+_65772443 | 3.45 |
ENSRNOT00000013296
|
Sebox
|
SEBOX homeobox |
| chr15_+_33822730 | 3.42 |
ENSRNOT00000067607
|
RGD1564324
|
similar to dehydrogenase/reductase member 2 |
| chr1_-_170471076 | 3.39 |
ENSRNOT00000025159
|
Arfip2
|
ADP-ribosylation factor interacting protein 2 |
| chr5_-_126096068 | 3.36 |
ENSRNOT00000036793
|
AABR07073134.1
|
|
| chr8_+_131845696 | 3.35 |
ENSRNOT00000005440
|
Tcaim
|
T cell activation inhibitor, mitochondrial |
| chr7_+_38945836 | 3.34 |
ENSRNOT00000006455
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr11_-_24294179 | 3.23 |
ENSRNOT00000002116
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr4_-_117126822 | 3.16 |
ENSRNOT00000086720
|
Rab11fip5
|
RAB11 family interacting protein 5 |
| chr10_+_84182118 | 3.12 |
ENSRNOT00000011027
|
Hoxb3
|
homeo box B3 |
| chr19_+_25029037 | 3.10 |
ENSRNOT00000045699
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr1_+_172822544 | 3.09 |
ENSRNOT00000090018
|
AABR07004960.1
|
|
| chr3_+_9643047 | 3.06 |
ENSRNOT00000035805
|
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr19_-_53497006 | 3.03 |
ENSRNOT00000074005
|
Fbxo31
|
F-box protein 31 |
| chr10_-_18574710 | 3.02 |
ENSRNOT00000079031
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr19_+_24496569 | 3.00 |
ENSRNOT00000004982
|
Mgat4d
|
MGAT4 family, member D |
| chrX_-_15428518 | 3.00 |
ENSRNOT00000075774
|
LOC108348172
|
proSAAS |
| chr14_+_42714315 | 2.97 |
ENSRNOT00000084095
ENSRNOT00000091449 |
Phox2b
|
paired-like homeobox 2b |
| chr5_+_135562034 | 2.95 |
ENSRNOT00000056967
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr16_-_48437223 | 2.94 |
ENSRNOT00000013005
ENSRNOT00000059401 |
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr1_+_72810545 | 2.92 |
ENSRNOT00000092117
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr13_+_42220251 | 2.92 |
ENSRNOT00000078185
|
AABR07020815.1
|
|
| chr16_+_20672374 | 2.90 |
ENSRNOT00000063780
|
RGD1566239
|
similar to RIKEN cDNA 2810428I15 |
| chr12_+_49419582 | 2.89 |
ENSRNOT00000082998
|
RGD1306556
|
similar to hypothetical protein A530094D01 |
| chr12_+_40466495 | 2.87 |
ENSRNOT00000001816
|
Aldh2
|
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr13_+_49092306 | 2.86 |
ENSRNOT00000000039
|
Nuak2
|
NUAK family kinase 2 |
| chr10_+_13196946 | 2.84 |
ENSRNOT00000071525
|
Prss32
|
protease, serine, 32 |
| chr8_+_20728817 | 2.83 |
ENSRNOT00000050101
|
Olr1179
|
olfactory receptor 1179 |
| chr9_-_11114843 | 2.78 |
ENSRNOT00000072031
|
Ebi3
|
Epstein-Barr virus induced 3 |
| chr1_+_102258124 | 2.74 |
ENSRNOT00000068434
|
Otog
|
otogelin |
| chrX_-_15327684 | 2.74 |
ENSRNOT00000009794
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr14_-_3462629 | 2.73 |
ENSRNOT00000061538
|
Brdt
|
bromodomain testis associated |
| chr3_-_177095597 | 2.71 |
ENSRNOT00000029386
|
Samd10
|
sterile alpha motif domain containing 10 |
| chr5_+_58393603 | 2.70 |
ENSRNOT00000080082
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr4_+_71795768 | 2.69 |
ENSRNOT00000031921
|
Tas2r135
|
taste receptor, type 2, member 135 |
| chr10_-_61772250 | 2.66 |
ENSRNOT00000092338
ENSRNOT00000085394 ENSRNOT00000046110 |
Srr
|
serine racemase |
| chr3_-_4394428 | 2.64 |
ENSRNOT00000044775
|
Abo
|
ABO, alpha 1-3-N-acetylgalactosaminyltransferase and alpha 1-3-galactosyltransferase |
| chr1_+_79982639 | 2.62 |
ENSRNOT00000071916
|
Dmwd
|
dystrophia myotonica, WD repeat containing |
| chr1_-_206394346 | 2.62 |
ENSRNOT00000038122
|
LOC100302465
|
hypothetical LOC100302465 |
| chr1_-_87937516 | 2.60 |
ENSRNOT00000087522
|
Eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr16_-_81834945 | 2.59 |
ENSRNOT00000037806
|
F7
|
coagulation factor VII |
| chr13_-_90832138 | 2.58 |
ENSRNOT00000010930
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr5_-_138697641 | 2.58 |
ENSRNOT00000012062
|
Guca2b
|
guanylate cyclase activator 2B |
| chr4_+_114835064 | 2.56 |
ENSRNOT00000031964
|
LOC103692165
|
rhotekin |
| chr5_+_63781801 | 2.55 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr18_-_70924708 | 2.53 |
ENSRNOT00000025257
|
Lipg
|
lipase G, endothelial type |
| chr1_+_220243473 | 2.52 |
ENSRNOT00000084650
ENSRNOT00000027091 |
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr1_-_81163689 | 2.50 |
ENSRNOT00000088746
|
LOC102548695
|
zinc finger protein 45-like |
| chr3_-_5402666 | 2.47 |
ENSRNOT00000061276
|
Abo2
|
histo-blood group ABO system transferase 2 |
| chr3_-_176706896 | 2.45 |
ENSRNOT00000017337
|
Ptk6
|
protein tyrosine kinase 6 |
| chr10_+_44804631 | 2.44 |
ENSRNOT00000040720
|
Olr1460
|
olfactory receptor 1460 |
| chr18_+_61261418 | 2.44 |
ENSRNOT00000064250
|
Zfp532
|
zinc finger protein 532 |
| chr5_+_157327657 | 2.43 |
ENSRNOT00000022847
|
Pla2g2e
|
phospholipase A2, group IIE |
| chr1_+_88542035 | 2.40 |
ENSRNOT00000032973
|
Zfp420
|
zinc finger protein 420 |
| chr6_+_101532518 | 2.37 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr4_+_9981958 | 2.35 |
ENSRNOT00000015322
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr8_+_39687269 | 2.35 |
ENSRNOT00000012060
|
Tmem218
|
transmembrane protein 218 |
| chr1_+_172051215 | 2.34 |
ENSRNOT00000048370
|
Olr235
|
olfactory receptor 235 |
| chr14_+_3882398 | 2.34 |
ENSRNOT00000033794
|
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog |
| chr4_+_79846521 | 2.32 |
ENSRNOT00000029332
|
Mpp6
|
membrane palmitoylated protein 6 |
| chr7_+_54859104 | 2.30 |
ENSRNOT00000087341
|
Caps2
|
calcyphosine 2 |
| chr1_+_85485289 | 2.27 |
ENSRNOT00000026182
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr10_+_109966904 | 2.23 |
ENSRNOT00000085821
|
Gps1
|
G protein pathway suppressor 1 |
| chr1_+_248402980 | 2.21 |
ENSRNOT00000043517
|
Chchd4
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr19_-_25345790 | 2.21 |
ENSRNOT00000010050
|
Zswim4
|
zinc finger, SWIM-type containing 4 |
| chr6_-_60124274 | 2.18 |
ENSRNOT00000059823
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr1_-_81559685 | 2.17 |
ENSRNOT00000075282
|
LOC100909620
|
CD177 antigen-like |
| chr8_-_55171718 | 2.16 |
ENSRNOT00000080736
|
LOC689959
|
hypothetical protein LOC689959 |
| chr4_+_174181644 | 2.14 |
ENSRNOT00000011555
|
Capza3
|
capping actin protein of muscle Z-line alpha subunit 3 |
| chr12_+_8032809 | 2.13 |
ENSRNOT00000082850
|
Slc7a1
|
solute carrier family 7 member 1 |
| chr4_+_1601652 | 2.13 |
ENSRNOT00000072284
|
LOC100912152
|
olfactory receptor 8B3-like |
| chr7_-_107774397 | 2.13 |
ENSRNOT00000093322
|
Ndrg1
|
N-myc downstream regulated 1 |
| chr18_+_63016577 | 2.11 |
ENSRNOT00000025147
|
Impa2
|
inositol monophosphatase 2 |
| chr13_+_71656651 | 2.11 |
ENSRNOT00000050365
|
Zfp648
|
zinc finger protein 648 |
| chr1_-_81152384 | 2.10 |
ENSRNOT00000026263
ENSRNOT00000077305 |
Zfp94
|
zinc finger protein 94 |
| chr5_-_152458023 | 2.08 |
ENSRNOT00000031225
|
Cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr8_+_42409187 | 2.08 |
ENSRNOT00000072605
|
Olr1256
|
olfactory receptor 1256 |
| chr10_-_102227554 | 2.08 |
ENSRNOT00000078035
|
AC096468.1
|
|
| chr1_-_264975132 | 2.06 |
ENSRNOT00000021748
|
Lbx1
|
ladybird homeobox 1 |
| chr8_-_82533689 | 2.04 |
ENSRNOT00000014124
|
Tmod2
|
tropomodulin 2 |
| chr6_+_101865917 | 2.04 |
ENSRNOT00000029888
|
Fam71d
|
family with sequence similarity 71, member D |
| chr6_+_28323647 | 2.02 |
ENSRNOT00000089093
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr6_-_3444519 | 2.00 |
ENSRNOT00000010366
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chrX_+_110789269 | 1.98 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_148240504 | 1.98 |
ENSRNOT00000085373
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr9_-_82336806 | 1.96 |
ENSRNOT00000024667
|
Slc23a3
|
solute carrier family 23, member 3 |
| chr6_+_107531528 | 1.94 |
ENSRNOT00000077555
|
Acot3
|
acyl-CoA thioesterase 3 |
| chr15_-_49467174 | 1.93 |
ENSRNOT00000019209
|
Adam7
|
ADAM metallopeptidase domain 7 |
| chr4_+_123586231 | 1.93 |
ENSRNOT00000049657
ENSRNOT00000013232 |
Grip2
|
glutamate receptor interacting protein 2 |
| chr15_+_120372 | 1.92 |
ENSRNOT00000007828
|
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr1_+_276309927 | 1.91 |
ENSRNOT00000067460
ENSRNOT00000066236 |
Vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr17_-_10559627 | 1.91 |
ENSRNOT00000023515
|
Higd2a
|
HIG1 hypoxia inducible domain family, member 2A |
| chr3_+_161433410 | 1.90 |
ENSRNOT00000024657
|
Slc12a5
|
solute carrier family 12 member 5 |
| chr1_+_79989019 | 1.89 |
ENSRNOT00000020428
|
Dmpk
|
dystrophia myotonica-protein kinase |
| chr10_-_87232723 | 1.87 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr20_-_4280795 | 1.86 |
ENSRNOT00000081431
|
RGD1624210
|
similar to butyrophilin-like 7 |
| chr12_-_50213400 | 1.85 |
ENSRNOT00000048541
|
LOC102552378
|
trichohyalin-like |
| chr12_+_52359310 | 1.85 |
ENSRNOT00000071857
ENSRNOT00000065893 |
Fbrsl1
|
fibrosin-like 1 |
| chr7_-_3074359 | 1.85 |
ENSRNOT00000019738
|
Ikzf4
|
IKAROS family zinc finger 4 |
| chr8_+_36625733 | 1.84 |
ENSRNOT00000016248
|
Cdon
|
cell adhesion associated, oncogene regulated |
| chr5_-_66322567 | 1.82 |
ENSRNOT00000032726
|
Cylc2
|
cylicin 2 |
| chr9_+_112360419 | 1.82 |
ENSRNOT00000086682
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr7_+_54859326 | 1.81 |
ENSRNOT00000039141
|
Caps2
|
calcyphosine 2 |
| chr5_-_168004724 | 1.81 |
ENSRNOT00000024711
|
Park7
|
Parkinsonism associated deglycase |
| chr16_-_20640720 | 1.80 |
ENSRNOT00000083254
|
AABR07024877.1
|
|
| chr4_+_145399913 | 1.79 |
ENSRNOT00000012177
|
Jagn1
|
jagunal homolog 1 |
| chr5_-_150704117 | 1.78 |
ENSRNOT00000067455
|
Sesn2
|
sestrin 2 |
| chr13_+_99005142 | 1.78 |
ENSRNOT00000004293
|
Acbd3
|
acyl-CoA binding domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx15
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 3.0 | 3.0 | GO:0048880 | sensory system development(GO:0048880) |
| 2.5 | 7.4 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 2.4 | 19.6 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 2.1 | 16.6 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 2.0 | 8.1 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 1.9 | 5.7 | GO:0030091 | protein repair(GO:0030091) |
| 1.9 | 7.6 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.9 | 5.7 | GO:0046439 | cysteine catabolic process(GO:0009093) glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) oxalic acid secretion(GO:0046724) |
| 1.8 | 10.7 | GO:0031427 | response to methotrexate(GO:0031427) |
| 1.8 | 21.0 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 1.5 | 10.8 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.4 | 4.3 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 1.4 | 6.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.3 | 6.5 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 1.2 | 9.5 | GO:0042904 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.2 | 10.5 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 1.0 | 3.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) peptidyl-glycine modification(GO:0018201) |
| 1.0 | 10.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 1.0 | 6.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 1.0 | 4.8 | GO:0002018 | angiotensin maturation(GO:0002003) renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.9 | 9.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.9 | 2.7 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.9 | 2.7 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.9 | 8.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.9 | 2.6 | GO:1905225 | response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) response to thyrotropin-releasing hormone(GO:1905225) |
| 0.8 | 2.5 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.8 | 12.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.8 | 5.5 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.8 | 3.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.8 | 9.3 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.7 | 2.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.7 | 5.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.6 | 1.9 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.6 | 3.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.6 | 68.5 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.6 | 1.8 | GO:0050787 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) TRAIL receptor biosynthetic process(GO:0045557) regulation of TRAIL receptor biosynthetic process(GO:0045560) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.6 | 5.4 | GO:0072338 | allantoin metabolic process(GO:0000255) isoleucine metabolic process(GO:0006549) creatine metabolic process(GO:0006600) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.6 | 2.4 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.6 | 1.7 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.6 | 2.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.5 | 2.6 | GO:0051691 | cellular oligosaccharide metabolic process(GO:0051691) |
| 0.5 | 12.4 | GO:0046852 | negative regulation of bone mineralization(GO:0030502) positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.5 | 1.6 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.5 | 8.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.5 | 3.6 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.5 | 1.5 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.5 | 3.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.5 | 1.5 | GO:2000040 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.5 | 5.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.5 | 1.9 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.5 | 6.6 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.5 | 2.4 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.5 | 5.7 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.4 | 1.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.4 | 3.9 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.4 | 1.7 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.4 | 1.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.4 | 1.9 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.4 | 4.8 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.4 | 1.1 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.4 | 9.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 4.2 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) |
| 0.3 | 2.0 | GO:0044027 | hypermethylation of CpG island(GO:0044027) C-5 methylation of cytosine(GO:0090116) |
| 0.3 | 4.0 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.3 | 2.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) basic amino acid transmembrane transport(GO:1990822) |
| 0.3 | 3.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 2.9 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.3 | 2.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 7.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) male genitalia development(GO:0030539) |
| 0.3 | 2.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.3 | 1.8 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.3 | 2.9 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.3 | 0.5 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.3 | 3.0 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.3 | 0.5 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.2 | 2.7 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.2 | 1.2 | GO:0044704 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) |
| 0.2 | 1.9 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 1.7 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 1.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 2.6 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 0.9 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 0.2 | 0.7 | GO:2000853 | negative regulation of corticosterone secretion(GO:2000853) |
| 0.2 | 0.9 | GO:0045226 | hyaluranon cable assembly(GO:0036118) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 | 2.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 16.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 0.6 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 1.0 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.2 | 2.7 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.2 | 3.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.2 | 2.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 1.9 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 8.7 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.2 | 2.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 2.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 1.8 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.2 | 0.5 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 2.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 2.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.4 | GO:2000541 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 0.1 | 2.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 2.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 2.6 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 3.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.9 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.4 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 2.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 3.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 1.7 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.1 | 1.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 2.3 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 4.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.6 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 0.1 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.1 | 2.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 2.2 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 5.6 | GO:0048477 | oogenesis(GO:0048477) |
| 0.1 | 0.6 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.1 | 0.7 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.5 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.1 | 0.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 5.2 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.1 | 1.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.4 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 1.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 2.7 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 2.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 7.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 1.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 1.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 3.2 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.2 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 22.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 2.1 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 2.6 | GO:0007018 | microtubule-based movement(GO:0007018) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.7 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 1.1 | 5.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.8 | 4.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.7 | 8.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.7 | 4.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.4 | 0.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.3 | 4.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 2.6 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.3 | 0.9 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.3 | 4.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 5.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 3.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.2 | 3.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 1.8 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 2.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 1.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 10.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 1.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 7.9 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 5.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 8.1 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 1.6 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 15.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 3.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 9.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 7.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.5 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 12.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.1 | GO:0044615 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.7 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 6.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 2.0 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 10.7 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 4.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 4.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 9.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 6.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 5.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 4.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.9 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 7.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 2.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 0.3 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 4.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 2.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 11.6 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 3.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 3.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 13.1 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 29.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 2.2 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 1.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 8.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 70.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 3.6 | 10.7 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 3.5 | 10.5 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 3.2 | 9.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 3.1 | 12.3 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 2.4 | 19.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 2.3 | 6.9 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 2.1 | 10.6 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 1.7 | 8.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 1.5 | 12.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 1.4 | 8.7 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 1.4 | 5.7 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 1.4 | 5.7 | GO:0016160 | amylase activity(GO:0016160) |
| 1.4 | 4.1 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 1.4 | 5.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 1.3 | 10.7 | GO:0031404 | chloride ion binding(GO:0031404) |
| 1.1 | 5.7 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 1.0 | 3.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.8 | 4.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.8 | 6.5 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.8 | 5.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.7 | 2.9 | GO:0070404 | NADH binding(GO:0070404) |
| 0.7 | 8.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.6 | 7.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.6 | 1.8 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.6 | 2.4 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.5 | 2.7 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.5 | 9.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.5 | 2.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.5 | 5.7 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.5 | 4.2 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.5 | 5.5 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.4 | 3.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 1.3 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.4 | 2.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.4 | 10.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.4 | 2.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.4 | 1.7 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.4 | 2.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 2.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.4 | 8.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.4 | 2.6 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.4 | 2.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.4 | 1.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.3 | 1.0 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.3 | 3.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 2.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 0.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.3 | 2.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) |
| 0.3 | 1.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 9.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 10.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 4.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 3.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 5.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 3.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 0.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.2 | 3.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 0.6 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 6.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 1.9 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 3.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 4.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 2.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 2.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 2.6 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 1.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.9 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.1 | 1.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 16.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 2.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.7 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 3.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.5 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 2.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 3.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 3.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.9 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 2.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 2.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 5.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 7.0 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.1 | 4.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 2.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 2.0 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 7.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 12.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 1.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 3.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 8.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 3.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 2.7 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 3.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 3.3 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 5.6 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 2.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 2.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.7 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 3.7 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 22.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 4.2 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 5.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 5.4 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.4 | GO:0015485 | cholesterol binding(GO:0015485) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 4.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 8.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 1.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 4.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 5.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 3.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 0.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 4.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 7.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 3.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 21.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 1.9 | 66.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 1.5 | 12.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.9 | 5.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.9 | 10.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.5 | 9.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.3 | 2.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 5.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 3.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 3.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 6.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 4.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 8.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 3.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 3.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 2.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 4.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 7.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.8 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.7 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 2.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 2.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 4.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.0 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |