Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
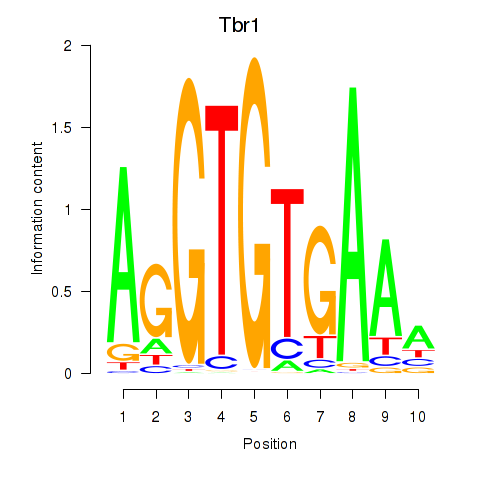
Results for Tbr1
Z-value: 1.00
Transcription factors associated with Tbr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbr1
|
ENSRNOG00000005049 | T-box, brain, 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbr1 | rn6_v1_chr3_+_47677720_47677720 | -0.14 | 1.3e-02 | Click! |
Activity profile of Tbr1 motif
Sorted Z-values of Tbr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_74057361 | 41.06 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chrX_-_1848904 | 37.76 |
ENSRNOT00000010984
|
Rgn
|
regucalcin |
| chr1_+_48273611 | 35.49 |
ENSRNOT00000022254
ENSRNOT00000022068 |
Slc22a1
|
solute carrier family 22 member 1 |
| chr2_-_23289266 | 32.56 |
ENSRNOT00000061708
|
Bhmt2
|
betaine-homocysteine S-methyltransferase 2 |
| chr9_+_4072823 | 31.96 |
ENSRNOT00000085330
ENSRNOT00000061864 |
LOC100910057
|
sulfotransferase 1C2-like |
| chr1_+_148240504 | 31.64 |
ENSRNOT00000085373
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr7_+_34326087 | 30.96 |
ENSRNOT00000006971
|
Hal
|
histidine ammonia lyase |
| chr15_+_57290849 | 30.61 |
ENSRNOT00000014909
|
Cpb2
|
carboxypeptidase B2 |
| chr11_-_81444375 | 30.11 |
ENSRNOT00000058479
ENSRNOT00000078131 ENSRNOT00000080949 ENSRNOT00000080562 ENSRNOT00000084867 |
Kng1
|
kininogen 1 |
| chr9_-_4683160 | 28.45 |
ENSRNOT00000081413
|
Sult1c2
|
sulfotransferase family 1C member 2 |
| chr12_+_19196611 | 27.89 |
ENSRNOT00000001801
|
Azgp1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr16_-_32868680 | 27.63 |
ENSRNOT00000015974
ENSRNOT00000082392 |
Aadat
|
aminoadipate aminotransferase |
| chr8_-_130550388 | 25.62 |
ENSRNOT00000026355
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr14_-_19072677 | 24.45 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr4_+_154215250 | 24.29 |
ENSRNOT00000072465
|
Mug2
|
murinoglobulin 2 |
| chr8_-_50531423 | 24.00 |
ENSRNOT00000090985
ENSRNOT00000074942 |
Apoc3
|
apolipoprotein C3 |
| chr16_-_7026540 | 23.00 |
ENSRNOT00000051751
|
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr19_-_41686229 | 22.79 |
ENSRNOT00000022721
|
Tat
|
tyrosine aminotransferase |
| chr13_-_80819218 | 22.05 |
ENSRNOT00000072922
|
Fmo6
|
flavin containing monooxygenase 6 |
| chr13_+_47572219 | 21.67 |
ENSRNOT00000088449
ENSRNOT00000087664 ENSRNOT00000005853 |
Pigr
|
polymeric immunoglobulin receptor |
| chr6_-_127653124 | 21.66 |
ENSRNOT00000047324
|
Serpina11
|
serpin family A member 11 |
| chr1_-_258875572 | 21.65 |
ENSRNOT00000093005
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr3_+_142383278 | 21.60 |
ENSRNOT00000017742
|
Foxa2
|
forkhead box A2 |
| chr6_-_129010271 | 20.84 |
ENSRNOT00000075378
|
Serpina10
|
serpin family A member 10 |
| chr14_+_22142364 | 20.41 |
ENSRNOT00000002699
|
Sult1b1
|
sulfotransferase family 1B member 1 |
| chr6_-_127620296 | 20.25 |
ENSRNOT00000012577
|
Serpina1
|
serpin family A member 1 |
| chr6_-_7058314 | 19.98 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr1_-_219312240 | 19.95 |
ENSRNOT00000066691
|
RGD1307603
|
similar to hypothetical protein MGC37914 |
| chr7_-_138483612 | 19.51 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr2_+_252452269 | 19.38 |
ENSRNOT00000021970
|
Uox
|
urate oxidase |
| chrX_-_13601069 | 19.32 |
ENSRNOT00000004686
|
Otc
|
ornithine carbamoyltransferase |
| chr2_-_53185926 | 19.27 |
ENSRNOT00000081558
|
Ghr
|
growth hormone receptor |
| chr1_+_21613148 | 19.21 |
ENSRNOT00000018695
|
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr10_+_109707962 | 19.18 |
ENSRNOT00000054962
ENSRNOT00000088898 |
Gcgr
|
glucagon receptor |
| chr20_-_45126062 | 18.81 |
ENSRNOT00000000720
|
RGD1310495
|
similar to KIAA1919 protein |
| chr9_+_100281339 | 18.38 |
ENSRNOT00000029127
|
Agxt
|
alanine-glyoxylate aminotransferase |
| chr16_-_81834945 | 17.65 |
ENSRNOT00000037806
|
F7
|
coagulation factor VII |
| chr1_-_48559162 | 17.10 |
ENSRNOT00000080352
|
Plg
|
plasminogen |
| chr9_-_26707571 | 17.06 |
ENSRNOT00000080948
|
AABR07067023.1
|
|
| chr1_-_102780381 | 16.93 |
ENSRNOT00000080132
|
Saa4
|
serum amyloid A4 |
| chr9_+_95221474 | 16.92 |
ENSRNOT00000066839
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr5_-_77408323 | 16.79 |
ENSRNOT00000046857
ENSRNOT00000046760 |
LOC259245
Mup4
|
alpha-2u globulin PGCL5 major urinary protein 4 |
| chr2_-_211322719 | 16.71 |
ENSRNOT00000027493
|
RGD1310209
|
similar to KIAA1324 protein |
| chr9_-_4685383 | 16.53 |
ENSRNOT00000061896
|
Sult1c2
|
sulfotransferase family 1C member 2 |
| chr5_-_77433847 | 16.19 |
ENSRNOT00000076906
ENSRNOT00000043056 |
LOC500473
Mup4
|
similar to alpha-2u globulin PGCL2 major urinary protein 4 |
| chr1_-_259287684 | 16.00 |
ENSRNOT00000054724
|
Cyp2c22
|
cytochrome P450, family 2, subfamily c, polypeptide 22 |
| chr13_+_99335020 | 15.85 |
ENSRNOT00000029787
|
AABR07021930.1
|
|
| chr13_-_80703615 | 15.42 |
ENSRNOT00000004599
|
Fmo4
|
flavin containing monooxygenase 4 |
| chr5_+_60528997 | 15.41 |
ENSRNOT00000051445
|
Grhpr
|
glyoxylate and hydroxypyruvate reductase |
| chr20_-_29029905 | 15.41 |
ENSRNOT00000075682
|
Oit3
|
oncoprotein induced transcript 3 |
| chr14_+_22937421 | 15.16 |
ENSRNOT00000065079
|
RGD1559459
|
similar to Expressed sequence AI788959 |
| chr5_-_77342299 | 15.04 |
ENSRNOT00000075994
|
RGD1566134
|
similar to alpha-2u-globulin |
| chr4_+_122244711 | 14.97 |
ENSRNOT00000038251
|
Uroc1
|
urocanate hydratase 1 |
| chr5_-_77316764 | 14.89 |
ENSRNOT00000071395
ENSRNOT00000076464 |
Mup4
|
major urinary protein 4 |
| chr14_+_22724070 | 14.85 |
ENSRNOT00000089471
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr9_+_4817854 | 14.79 |
ENSRNOT00000040879
|
LOC100910526
|
sulfotransferase 1C2-like |
| chr16_-_7758189 | 14.72 |
ENSRNOT00000026588
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr1_-_219311502 | 14.70 |
ENSRNOT00000085439
|
RGD1307603
|
similar to hypothetical protein MGC37914 |
| chr9_+_4807717 | 14.58 |
ENSRNOT00000092159
|
LOC100910526
|
sulfotransferase 1C2-like |
| chr5_-_52767592 | 14.57 |
ENSRNOT00000048383
|
LOC298111
|
alpha2u globulin |
| chr2_-_210766501 | 14.46 |
ENSRNOT00000025800
|
Gstm3
|
glutathione S-transferase mu 3 |
| chr1_-_166037424 | 14.30 |
ENSRNOT00000026115
|
P2ry2
|
purinergic receptor P2Y2 |
| chr8_+_116857684 | 14.29 |
ENSRNOT00000026711
|
Mst1
|
macrophage stimulating 1 |
| chr1_+_40086470 | 14.22 |
ENSRNOT00000021895
|
Iyd
|
iodotyrosine deiodinase |
| chr9_+_81968332 | 14.17 |
ENSRNOT00000023152
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr2_+_54466280 | 14.01 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr17_+_8489266 | 13.98 |
ENSRNOT00000016252
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr5_+_61425746 | 13.86 |
ENSRNOT00000064113
|
RGD1305807
|
hypothetical LOC298077 |
| chr1_+_282568287 | 13.76 |
ENSRNOT00000015997
|
Ces2i
|
carboxylesterase 2I |
| chr5_-_77725867 | 13.72 |
ENSRNOT00000076791
|
Rn50_5_0814.4
|
alpha2u globulin (LOC298111), mRNA |
| chr3_-_2727616 | 13.60 |
ENSRNOT00000061904
|
C8g
|
complement C8 gamma chain |
| chr2_-_210749991 | 13.57 |
ENSRNOT00000051261
ENSRNOT00000052403 |
Gstm6l
|
glutathione S-transferase, mu 6-like |
| chr1_+_64506735 | 13.16 |
ENSRNOT00000086331
|
Nlrp12
|
NLR family, pyrin domain containing 12 |
| chr9_+_16825845 | 13.13 |
ENSRNOT00000074821
|
LOC100910410
|
ly6/PLAUR domain-containing protein 4-like |
| chrX_+_62282212 | 12.98 |
ENSRNOT00000039568
|
AABR07038837.1
|
|
| chr3_-_176706896 | 12.96 |
ENSRNOT00000017337
|
Ptk6
|
protein tyrosine kinase 6 |
| chr5_-_77903062 | 12.79 |
ENSRNOT00000073954
ENSRNOT00000059320 ENSRNOT00000089382 ENSRNOT00000076074 ENSRNOT00000074349 |
LOC100912565
Rn50_5_0814.4
|
major urinary protein-like alpha2u globulin (LOC298111), mRNA |
| chr3_+_95233874 | 12.72 |
ENSRNOT00000079743
|
LOC691083
|
hypothetical protein LOC691083 |
| chr4_-_123557501 | 12.58 |
ENSRNOT00000075042
ENSRNOT00000085966 |
Aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr7_+_14891001 | 12.34 |
ENSRNOT00000041034
|
Cyp4f4
|
cytochrome P450, family 4, subfamily f, polypeptide 4 |
| chr14_-_80973456 | 12.21 |
ENSRNOT00000013257
|
Hgfac
|
HGF activator |
| chr5_-_77492013 | 12.20 |
ENSRNOT00000012293
|
LOC259245
|
alpha-2u globulin PGCL5 |
| chrX_+_6430594 | 11.94 |
ENSRNOT00000044009
|
Maob
|
monoamine oxidase B |
| chr10_-_50402616 | 11.93 |
ENSRNOT00000004546
|
Hs3st3b1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
| chr2_-_210738378 | 11.91 |
ENSRNOT00000025746
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr10_-_71491743 | 11.82 |
ENSRNOT00000038955
|
LOC102552988
|
uncharacterized LOC102552988 |
| chr2_-_195423787 | 11.57 |
ENSRNOT00000071603
|
LOC103689947
|
selenium-binding protein 1 |
| chr8_-_21901829 | 11.55 |
ENSRNOT00000027963
|
Angptl6
|
angiopoietin-like 6 |
| chr19_+_24496569 | 11.46 |
ENSRNOT00000004982
|
Mgat4d
|
MGAT4 family, member D |
| chr9_-_10182676 | 11.45 |
ENSRNOT00000074894
|
LOC316124
|
similar to gonadotropin-regulated long chain acyl-CoA synthetase |
| chr13_+_75105615 | 11.41 |
ENSRNOT00000076653
|
Tp53i3
|
tumor protein p53 inducible protein 3 |
| chr14_-_2934571 | 11.37 |
ENSRNOT00000049772
|
Ube2d4
|
ubiquitin-conjugating enzyme E2D 4 (putative) |
| chr5_+_137357674 | 11.29 |
ENSRNOT00000092813
|
RGD1305347
|
similar to RIKEN cDNA 2610528J11 |
| chr2_+_20857202 | 11.28 |
ENSRNOT00000078919
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr12_-_41627741 | 11.12 |
ENSRNOT00000001875
|
Sds
|
serine dehydratase |
| chr1_+_144069638 | 11.07 |
ENSRNOT00000026870
|
Sh3gl3
|
SH3 domain-containing GRB2-like 3 |
| chr11_-_80936524 | 10.84 |
ENSRNOT00000044591
|
St6gal1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr2_+_127489771 | 10.83 |
ENSRNOT00000093581
|
Intu
|
inturned planar cell polarity protein |
| chr10_-_70118194 | 10.74 |
ENSRNOT00000009944
|
Cct6b
|
chaperonin containing TCP1 subunit 6B |
| chr2_-_196113149 | 10.69 |
ENSRNOT00000088465
|
Selenbp1
|
selenium binding protein 1 |
| chr8_-_68525911 | 10.67 |
ENSRNOT00000080588
ENSRNOT00000011109 |
Iqch
|
IQ motif containing H |
| chr2_-_158133861 | 10.66 |
ENSRNOT00000090700
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr18_+_69841053 | 10.60 |
ENSRNOT00000071545
ENSRNOT00000030613 ENSRNOT00000075543 |
Mro
|
maestro |
| chr1_-_20155960 | 10.50 |
ENSRNOT00000061389
|
Samd3
|
sterile alpha motif domain containing 3 |
| chr2_-_181900856 | 10.43 |
ENSRNOT00000082156
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr5_-_115222075 | 10.33 |
ENSRNOT00000058163
ENSRNOT00000012503 |
LOC100912642
|
cytochrome P450 2J3-like |
| chr9_+_95233957 | 10.27 |
ENSRNOT00000071003
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr7_+_54859104 | 10.25 |
ENSRNOT00000087341
|
Caps2
|
calcyphosine 2 |
| chr15_+_23777856 | 10.10 |
ENSRNOT00000060847
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr9_-_95290931 | 10.03 |
ENSRNOT00000025300
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3 |
| chr11_+_83883879 | 9.88 |
ENSRNOT00000050927
|
LOC108348101
|
chloride channel protein 2 |
| chr6_+_110749705 | 9.85 |
ENSRNOT00000084348
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr20_+_40769586 | 9.82 |
ENSRNOT00000001079
|
Fabp7
|
fatty acid binding protein 7 |
| chr10_-_109747987 | 9.81 |
ENSRNOT00000054958
|
P4hb
|
prolyl 4-hydroxylase subunit beta |
| chr1_+_72810545 | 9.71 |
ENSRNOT00000092117
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr1_-_199655147 | 9.67 |
ENSRNOT00000026979
|
LOC103691238
|
zinc finger protein 239-like |
| chr1_+_222310920 | 9.64 |
ENSRNOT00000091465
|
Macrod1
|
MACRO domain containing 1 |
| chr4_+_153921115 | 9.62 |
ENSRNOT00000018821
|
Slc6a12
|
solute carrier family 6 member 12 |
| chr1_+_166428761 | 9.60 |
ENSRNOT00000085555
|
Stard10
|
StAR-related lipid transfer domain containing 10 |
| chrX_-_65335987 | 9.43 |
ENSRNOT00000047128
|
AABR07038981.1
|
|
| chr3_-_5802129 | 9.31 |
ENSRNOT00000009555
|
Sardh
|
sarcosine dehydrogenase |
| chr2_-_210703265 | 9.16 |
ENSRNOT00000080228
|
Gstm6l
|
glutathione S-transferase, mu 6-like |
| chr6_+_111180108 | 9.00 |
ENSRNOT00000082027
|
Gstz1
|
glutathione S-transferase zeta 1 |
| chr3_+_54734015 | 8.85 |
ENSRNOT00000034341
|
LOC685184
|
similar to protein expressed in prostate, ovary, testis, and placenta 8 isoform 2 |
| chr18_+_46148849 | 8.68 |
ENSRNOT00000026724
|
Prr16
|
proline rich 16 |
| chr16_-_24951612 | 8.68 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chr13_-_37287458 | 8.65 |
ENSRNOT00000003391
|
Insig2
|
insulin induced gene 2 |
| chr8_-_103608913 | 8.61 |
ENSRNOT00000013209
|
Pls1
|
plastin 1 |
| chr2_-_19808937 | 8.60 |
ENSRNOT00000044237
|
Atp6ap1l
|
ATPase H+ transporting accessory protein 1 like |
| chr8_+_111495331 | 8.59 |
ENSRNOT00000091275
ENSRNOT00000012162 |
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chrX_+_110818716 | 8.56 |
ENSRNOT00000086308
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr6_+_107517668 | 8.53 |
ENSRNOT00000013753
|
Acot4
|
acyl-CoA thioesterase 4 |
| chr5_+_162127810 | 8.51 |
ENSRNOT00000038858
|
Pramef27
|
PRAME family member 27 |
| chr5_+_75392790 | 8.49 |
ENSRNOT00000048457
|
Musk
|
muscle associated receptor tyrosine kinase |
| chr8_-_78233430 | 8.47 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr10_+_110346453 | 8.46 |
ENSRNOT00000054928
|
Tex19.1
|
testis expressed 19.1 |
| chr8_+_117068582 | 8.45 |
ENSRNOT00000073559
|
Amt
|
aminomethyltransferase |
| chr7_+_54859326 | 8.39 |
ENSRNOT00000039141
|
Caps2
|
calcyphosine 2 |
| chr5_-_173483825 | 8.32 |
ENSRNOT00000085087
|
Ttll10
|
tubulin tyrosine ligase like 10 |
| chr4_-_45414177 | 8.31 |
ENSRNOT00000091311
|
Asz1
|
ankyrin repeat, SAM and basic leucine zipper domain containing 1 |
| chr18_+_47613854 | 8.29 |
ENSRNOT00000025270
|
Zfp474
|
zinc finger protein 474 |
| chr11_+_88699222 | 8.27 |
ENSRNOT00000084177
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr6_+_36089433 | 8.25 |
ENSRNOT00000090438
ENSRNOT00000005731 |
Nt5c1b
|
5'-nucleotidase, cytosolic IB |
| chr4_+_163162211 | 8.13 |
ENSRNOT00000082537
|
Clec1b
|
C-type lectin domain family 1, member B |
| chr12_+_40466495 | 8.09 |
ENSRNOT00000001816
|
Aldh2
|
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr4_-_161907767 | 8.07 |
ENSRNOT00000009557
|
A2ml1
|
alpha-2-macroglobulin-like 1 |
| chr16_+_68633720 | 8.02 |
ENSRNOT00000081838
|
LOC100911229
|
sperm motility kinase-like |
| chr8_+_44001096 | 7.96 |
ENSRNOT00000084164
|
Tmem225
|
transmembrane protein 225 |
| chr7_+_77066955 | 7.94 |
ENSRNOT00000008700
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr5_+_133896141 | 7.92 |
ENSRNOT00000011434
|
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr19_-_860493 | 7.89 |
ENSRNOT00000078469
|
LOC102554842
|
CKLF-like MARVEL transmembrane domain-containing protein 2B-like |
| chr2_-_210703606 | 7.89 |
ENSRNOT00000077563
|
Gstm6l
|
glutathione S-transferase, mu 6-like |
| chr2_+_260039651 | 7.79 |
ENSRNOT00000073873
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr10_+_13196946 | 7.74 |
ENSRNOT00000071525
|
Prss32
|
protease, serine, 32 |
| chr14_-_82287706 | 7.66 |
ENSRNOT00000080695
|
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr10_+_103395511 | 7.65 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chrX_-_111102464 | 7.65 |
ENSRNOT00000084176
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr7_+_38945836 | 7.64 |
ENSRNOT00000006455
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr4_+_68849033 | 7.59 |
ENSRNOT00000016912
|
Mgam
|
maltase-glucoamylase |
| chr17_-_8607494 | 7.54 |
ENSRNOT00000016138
|
Slc25a48
|
solute carrier family 25, member 48 |
| chr11_+_16826399 | 7.52 |
ENSRNOT00000050701
|
Cxadr
|
coxsackie virus and adenovirus receptor |
| chr10_-_98294522 | 7.50 |
ENSRNOT00000005489
|
Abca8
|
ATP binding cassette subfamily A member 8 |
| chr11_+_82862695 | 7.45 |
ENSRNOT00000071707
ENSRNOT00000073435 |
Clcn2
|
chloride voltage-gated channel 2 |
| chr11_-_32508420 | 7.42 |
ENSRNOT00000002717
|
Kcne1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chr16_-_70006016 | 7.33 |
ENSRNOT00000092030
|
Gm1141
|
predicted gene 1141 |
| chr4_+_174181644 | 7.33 |
ENSRNOT00000011555
|
Capza3
|
capping actin protein of muscle Z-line alpha subunit 3 |
| chr9_+_81689802 | 7.31 |
ENSRNOT00000021432
|
Vil1
|
villin 1 |
| chr14_-_44613904 | 7.30 |
ENSRNOT00000003811
|
Klb
|
klotho beta |
| chr8_+_52829085 | 7.22 |
ENSRNOT00000007754
|
RGD1563941
|
similar to hypothetical protein FLJ20010 |
| chr4_-_85192834 | 7.19 |
ENSRNOT00000043752
|
Ggct
|
gamma-glutamyl cyclotransferase |
| chr2_-_252451999 | 7.18 |
ENSRNOT00000021861
|
Dnase2b
|
deoxyribonuclease 2 beta |
| chr5_-_58288125 | 7.16 |
ENSRNOT00000068752
|
Fam205a
|
family with sequence similarity 205, member A |
| chr14_-_92495894 | 7.15 |
ENSRNOT00000064483
|
Cobl
|
cordon-bleu WH2 repeat protein |
| chr18_+_16616937 | 7.15 |
ENSRNOT00000093641
|
Mocos
|
molybdenum cofactor sulfurase |
| chr2_+_84645084 | 7.14 |
ENSRNOT00000015448
|
Cmbl
|
carboxymethylenebutenolidase homolog |
| chr2_-_158156150 | 7.12 |
ENSRNOT00000016621
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr4_-_179904844 | 7.02 |
ENSRNOT00000085386
|
Lmntd1
|
lamin tail domain containing 1 |
| chr14_-_3462629 | 6.99 |
ENSRNOT00000061538
|
Brdt
|
bromodomain testis associated |
| chrX_+_16946207 | 6.98 |
ENSRNOT00000003981
|
RGD1563606
|
similar to cysteine-rich protein 2 |
| chr9_-_60330086 | 6.98 |
ENSRNOT00000092162
ENSRNOT00000093630 |
Dnah7
|
dynein, axonemal, heavy chain 7 |
| chr1_-_100205147 | 6.93 |
ENSRNOT00000046737
|
AABR07003250.1
|
|
| chr4_+_122365093 | 6.91 |
ENSRNOT00000024011
|
Klf15
|
Kruppel-like factor 15 |
| chr17_-_32076181 | 6.89 |
ENSRNOT00000074842
|
LOC100911107
|
leukocyte elastase inhibitor A-like |
| chr4_+_22084954 | 6.89 |
ENSRNOT00000090968
|
Crot
|
carnitine O-octanoyltransferase |
| chr1_+_152072665 | 6.88 |
ENSRNOT00000022538
|
Rab38
|
RAB38, member RAS oncogene family |
| chr1_-_247476827 | 6.81 |
ENSRNOT00000021298
|
Insl6
|
insulin-like 6 |
| chr9_+_64066579 | 6.79 |
ENSRNOT00000013511
|
RGD1306941
|
similar to CG31122-PA |
| chr3_+_143151739 | 6.79 |
ENSRNOT00000006850
|
Cst13
|
cystatin 13 |
| chr12_-_48238887 | 6.76 |
ENSRNOT00000078868
|
Acacb
|
acetyl-CoA carboxylase beta |
| chr2_+_93669765 | 6.74 |
ENSRNOT00000045438
|
Slc10a5
|
solute carrier family 10, member 5 |
| chr1_+_209237233 | 6.70 |
ENSRNOT00000021537
|
Mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr13_+_49092306 | 6.69 |
ENSRNOT00000000039
|
Nuak2
|
NUAK family kinase 2 |
| chr16_-_54628458 | 6.68 |
ENSRNOT00000042587
|
Adam24
|
ADAM metallopeptidase domain 24 |
| chr2_+_211880262 | 6.68 |
ENSRNOT00000038315
|
Slc25a54
|
solute carrier family 25, member 54 |
| chr14_-_21898284 | 6.68 |
ENSRNOT00000036314
|
Prr27
|
proline rich 27 |
| chr10_+_44804631 | 6.65 |
ENSRNOT00000040720
|
Olr1460
|
olfactory receptor 1460 |
| chr11_+_44089797 | 6.65 |
ENSRNOT00000060861
|
LOC108352322
|
ferritin heavy chain-like |
| chr4_+_153921900 | 6.65 |
ENSRNOT00000089482
|
Slc6a12
|
solute carrier family 6 member 12 |
| chr1_+_219964429 | 6.63 |
ENSRNOT00000088288
|
Sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr6_+_10483308 | 6.60 |
ENSRNOT00000074516
|
Tmem247
|
transmembrane protein 247 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.6 | 37.8 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 10.3 | 31.0 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 10.2 | 30.6 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 9.2 | 27.6 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 8.0 | 24.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 7.5 | 15.0 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 7.2 | 21.6 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 6.8 | 20.2 | GO:0033986 | response to methanol(GO:0033986) |
| 6.5 | 19.5 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 6.4 | 19.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 6.1 | 18.4 | GO:0046724 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) oxalic acid secretion(GO:0046724) |
| 5.9 | 17.6 | GO:1905225 | response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) response to thyrotropin-releasing hormone(GO:1905225) |
| 5.7 | 17.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 5.3 | 31.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 5.0 | 20.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 4.8 | 14.5 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 4.7 | 14.2 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 4.7 | 14.0 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 4.6 | 13.9 | GO:0046865 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 4.4 | 13.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) negative regulation of Toll signaling pathway(GO:0045751) |
| 4.2 | 12.6 | GO:0042560 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 4.1 | 32.6 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 4.0 | 40.0 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 3.9 | 19.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 3.9 | 61.7 | GO:0051923 | sulfation(GO:0051923) |
| 3.6 | 10.8 | GO:1990743 | protein sialylation(GO:1990743) |
| 3.5 | 10.6 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 3.4 | 20.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 3.2 | 12.7 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 3.0 | 82.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 3.0 | 27.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 2.8 | 11.4 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 2.8 | 8.5 | GO:2000541 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 2.8 | 11.1 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 2.8 | 8.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 2.8 | 11.0 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 2.6 | 5.2 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 2.5 | 12.7 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 2.3 | 9.3 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 2.3 | 4.6 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 2.2 | 9.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 2.2 | 6.7 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 2.2 | 6.6 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 2.1 | 8.4 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 2.1 | 6.2 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 2.0 | 6.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) response to butyrate(GO:1903544) |
| 2.0 | 14.2 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 2.0 | 3.9 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 1.9 | 19.4 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 1.9 | 19.2 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 1.9 | 17.2 | GO:0046449 | allantoin metabolic process(GO:0000255) isoleucine metabolic process(GO:0006549) creatine metabolic process(GO:0006600) creatinine metabolic process(GO:0046449) |
| 1.9 | 3.8 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 1.9 | 5.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.8 | 5.4 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 1.8 | 7.1 | GO:0090381 | regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 1.7 | 10.5 | GO:0070541 | response to vitamin B1(GO:0010266) response to platinum ion(GO:0070541) |
| 1.7 | 5.2 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of apoptotic cell clearance(GO:2000426) |
| 1.7 | 5.2 | GO:0014028 | notochord formation(GO:0014028) |
| 1.7 | 5.2 | GO:0016129 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 1.7 | 6.9 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 1.7 | 5.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 1.6 | 9.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 1.6 | 4.8 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 1.6 | 14.3 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 1.5 | 7.5 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 1.5 | 7.5 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 1.5 | 7.3 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 1.4 | 4.3 | GO:2000118 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 1.4 | 7.2 | GO:0048669 | somite specification(GO:0001757) collateral sprouting in absence of injury(GO:0048669) |
| 1.4 | 11.3 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 1.4 | 5.6 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 1.4 | 11.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.4 | 6.9 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 1.4 | 5.5 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 1.3 | 9.4 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 1.3 | 13.4 | GO:0032782 | bile acid secretion(GO:0032782) |
| 1.3 | 16.0 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 1.3 | 10.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 1.3 | 6.5 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 1.3 | 13.8 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 1.2 | 5.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 1.2 | 32.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 1.2 | 7.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 1.2 | 24.6 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 1.2 | 4.6 | GO:0051692 | cellular oligosaccharide catabolic process(GO:0051692) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 1.2 | 3.5 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 1.1 | 3.4 | GO:0030091 | protein repair(GO:0030091) |
| 1.1 | 5.4 | GO:0036233 | glycine import(GO:0036233) |
| 1.1 | 1.1 | GO:1900453 | negative regulation of long term synaptic depression(GO:1900453) |
| 1.1 | 2.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 1.1 | 3.2 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 1.1 | 3.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 1.0 | 15.4 | GO:0042737 | drug catabolic process(GO:0042737) |
| 1.0 | 3.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 1.0 | 7.1 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 1.0 | 6.9 | GO:0030035 | microspike assembly(GO:0030035) |
| 1.0 | 3.9 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.0 | 6.7 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.9 | 7.5 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.9 | 3.8 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.9 | 3.7 | GO:2001190 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.9 | 9.0 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.9 | 5.4 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.9 | 33.0 | GO:0042311 | vasodilation(GO:0042311) |
| 0.9 | 2.6 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.9 | 3.5 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.9 | 3.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.9 | 0.9 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.9 | 2.6 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.9 | 2.6 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.9 | 2.6 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.9 | 2.6 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.9 | 0.9 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.8 | 2.5 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.8 | 22.4 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.8 | 3.3 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.8 | 2.4 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.8 | 3.9 | GO:0007343 | egg activation(GO:0007343) |
| 0.8 | 8.7 | GO:0071501 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.8 | 4.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.8 | 3.0 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.8 | 2.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.8 | 1.5 | GO:0021874 | Wnt signaling pathway involved in forebrain neuroblast division(GO:0021874) |
| 0.7 | 2.2 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.7 | 0.7 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.7 | 1.5 | GO:0017143 | insecticide metabolic process(GO:0017143) |
| 0.7 | 1.5 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.7 | 2.2 | GO:0031427 | response to methotrexate(GO:0031427) |
| 0.7 | 3.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.7 | 2.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.7 | 3.5 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.7 | 2.7 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.7 | 2.0 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.7 | 6.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.7 | 2.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.7 | 2.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.7 | 3.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.7 | 20.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.6 | 7.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.6 | 3.2 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.6 | 5.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.6 | 1.3 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.6 | 9.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.6 | 2.5 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.6 | 2.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.6 | 7.4 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.6 | 1.2 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.6 | 3.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.6 | 1.8 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.6 | 5.5 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.6 | 2.4 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.6 | 1.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.6 | 2.9 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.6 | 1.8 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.6 | 9.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.6 | 8.5 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.6 | 5.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.6 | 2.8 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.5 | 3.8 | GO:0042148 | strand invasion(GO:0042148) |
| 0.5 | 4.9 | GO:0015816 | glycine transport(GO:0015816) |
| 0.5 | 7.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.5 | 6.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.5 | 5.9 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.5 | 5.9 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.5 | 0.5 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.5 | 2.1 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.5 | 14.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.5 | 4.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.5 | 6.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.5 | 2.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.5 | 6.7 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.5 | 13.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.5 | 1.5 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.5 | 2.5 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.5 | 7.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.5 | 1.9 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.5 | 6.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.5 | 1.4 | GO:0048749 | compound eye development(GO:0048749) |
| 0.5 | 11.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.5 | 2.8 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.5 | 1.4 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.5 | 1.4 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.5 | 7.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.4 | 0.4 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.4 | 1.3 | GO:0015803 | branched-chain amino acid transport(GO:0015803) histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.4 | 7.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.4 | 4.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.4 | 5.3 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.4 | 1.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.4 | 11.3 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.4 | 4.3 | GO:0072350 | tricarboxylic acid metabolic process(GO:0072350) |
| 0.4 | 0.9 | GO:0051795 | catagen(GO:0042637) regulation of catagen(GO:0051794) positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.4 | 3.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.4 | 0.8 | GO:0060935 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.4 | 2.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.4 | 6.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.4 | 2.9 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.4 | 17.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.4 | 7.0 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.4 | 2.4 | GO:0048840 | otolith development(GO:0048840) |
| 0.4 | 4.4 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.4 | 2.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.4 | 5.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.4 | 1.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.4 | 1.6 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.4 | 3.9 | GO:0006833 | water transport(GO:0006833) |
| 0.4 | 2.3 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.4 | 3.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.4 | 1.5 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.4 | 4.2 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.4 | 3.7 | GO:0071287 | cellular response to manganese ion(GO:0071287) |
| 0.4 | 1.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 4.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.4 | 1.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.4 | 1.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.4 | 4.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.4 | 2.8 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.4 | 2.5 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.3 | 2.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.3 | 2.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.3 | 3.0 | GO:0051177 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.3 | 2.6 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.3 | 4.0 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.3 | 1.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 0.7 | GO:0071442 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.3 | 13.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.3 | 11.8 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.3 | 0.9 | GO:0071680 | response to indole-3-methanol(GO:0071680) |
| 0.3 | 9.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.3 | 1.2 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 0.3 | 9.0 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.3 | 2.5 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.3 | 1.9 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.3 | 1.8 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.3 | 3.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.3 | 4.6 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.3 | 7.3 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.3 | 6.0 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.3 | 3.6 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.3 | 9.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.3 | 2.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.3 | 3.6 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.3 | 1.5 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 0.3 | 3.5 | GO:0033141 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.3 | 2.5 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.3 | 6.9 | GO:2000757 | negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.3 | 0.5 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.3 | 0.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.3 | 2.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.3 | 1.4 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.3 | 1.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.3 | 4.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.3 | 0.8 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.3 | 1.3 | GO:0045297 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) |
| 0.3 | 0.8 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.3 | 2.6 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) |
| 0.3 | 7.2 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.3 | 2.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 | 0.8 | GO:0035983 | meiotic chromosome movement towards spindle pole(GO:0016344) response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.3 | 3.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.3 | 1.3 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 1.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 3.7 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.2 | 2.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 0.7 | GO:0010034 | response to acetate(GO:0010034) |
| 0.2 | 5.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.2 | 5.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 8.7 | GO:0007140 | male meiosis(GO:0007140) |
| 0.2 | 15.4 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.2 | 0.7 | GO:0036476 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.2 | 4.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 2.3 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 0.2 | 4.5 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.2 | 3.1 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.2 | 0.4 | GO:0009946 | proximal/distal axis specification(GO:0009946) specification of axis polarity(GO:0065001) |
| 0.2 | 3.0 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 1.5 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.2 | 8.4 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.2 | 0.4 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.2 | 47.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.2 | 3.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.2 | 1.0 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.2 | 1.6 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.2 | 2.4 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 0.8 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.2 | 0.8 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 5.6 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.2 | 1.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 2.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 5.1 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.2 | 0.9 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.2 | 0.6 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 1.5 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.2 | 0.5 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.2 | 1.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 2.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.2 | 0.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 0.9 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.2 | 1.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.3 | GO:0015866 | ADP transport(GO:0015866) |
| 0.2 | 3.5 | GO:0019068 | virion assembly(GO:0019068) |
| 0.2 | 1.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 19.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 1.5 | GO:1903671 | negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.1 | 1.6 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 | 2.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 1.7 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 6.0 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 1.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.5 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 2.8 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 2.9 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.9 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.1 | 1.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 2.7 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 2.3 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 0.4 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.1 | 2.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 118.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.5 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 1.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 2.3 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.1 | 0.8 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 1.4 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 2.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.8 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 1.4 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 1.0 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.1 | 0.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 1.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 2.3 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 1.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 2.9 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 1.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 2.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 3.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 3.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.8 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.1 | 1.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.0 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 3.1 | GO:0006638 | neutral lipid metabolic process(GO:0006638) |
| 0.0 | 1.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.9 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 5.4 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.6 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 0.3 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 21.2 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.0 | 0.5 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.7 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.3 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.1 | GO:1990592 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:2000853 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) negative regulation of corticosterone secretion(GO:2000853) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.5 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 1.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.9 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 17.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 4.0 | 24.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 3.9 | 19.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 3.7 | 11.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 2.4 | 26.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 2.3 | 14.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 2.3 | 15.8 | GO:1990357 | terminal web(GO:1990357) |
| 1.9 | 38.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 1.7 | 7.0 | GO:0036156 | inner dynein arm(GO:0036156) |
| 1.4 | 12.2 | GO:0071546 | pi-body(GO:0071546) |
| 1.3 | 2.7 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 1.2 | 10.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 1.1 | 6.9 | GO:0031904 | endosome lumen(GO:0031904) |
| 1.1 | 5.6 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 1.1 | 4.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 1.0 | 19.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 1.0 | 5.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.0 | 14.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.0 | 18.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.9 | 22.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.8 | 5.5 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.8 | 90.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.8 | 10.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.7 | 9.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.7 | 0.7 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.7 | 8.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.7 | 3.5 | GO:0072557 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.7 | 5.3 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.7 | 2.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.6 | 2.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.6 | 1.8 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.6 | 3.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.6 | 3.3 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.6 | 2.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.5 | 6.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.5 | 5.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.5 | 3.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.5 | 9.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.5 | 4.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.5 | 3.3 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.5 | 1.4 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.4 | 1.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.4 | 59.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.4 | 4.2 | GO:0070187 | telosome(GO:0070187) |
| 0.4 | 1.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.4 | 2.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.4 | 53.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.4 | 19.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.4 | 1.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.4 | 3.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 16.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.4 | 2.5 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.3 | 2.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.3 | 1.5 | GO:1903768 | sweet taste receptor complex(GO:1903767) taste receptor complex(GO:1903768) |
| 0.3 | 2.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.3 | 2.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.3 | 0.8 | GO:0090534 | calcium ion-transporting ATPase complex(GO:0090534) |
| 0.3 | 2.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 2.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 1.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.3 | 1.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 2.3 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 3.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 12.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 0.7 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 1.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 2.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 10.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 12.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.2 | 3.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.2 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 1.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 0.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 5.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 54.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.2 | 2.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 4.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 4.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 3.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 3.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 6.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.0 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 10.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 13.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 3.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 41.3 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.1 | 8.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 3.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 145.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 1.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 29.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 1.3 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 0.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 10.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.9 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 51.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 6.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 0.5 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 1.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 72.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.5 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 1.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 3.6 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.6 | GO:0005903 | brush border(GO:0005903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.3 | 40.0 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 12.6 | 62.9 | GO:0005550 | pheromone binding(GO:0005550) |
| 9.7 | 48.7 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 9.2 | 27.6 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 8.0 | 24.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 7.6 | 22.8 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 6.4 | 19.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 4.9 | 39.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 4.8 | 19.3 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 4.8 | 19.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 3.8 | 15.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 3.7 | 11.2 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 3.7 | 18.4 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 3.5 | 10.5 | GO:0004655 | porphobilinogen synthase activity(GO:0004655) |
| 3.5 | 3.5 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 3.1 | 37.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 3.0 | 12.0 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 2.9 | 20.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 2.8 | 22.3 | GO:0008430 | selenium binding(GO:0008430) |
| 2.8 | 8.3 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 2.8 | 19.4 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 2.8 | 11.0 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 2.7 | 82.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 2.7 | 10.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 2.7 | 18.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 2.6 | 5.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 2.5 | 15.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 2.5 | 14.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 2.4 | 9.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 2.3 | 6.9 | GO:0016414 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 2.3 | 6.9 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 2.2 | 86.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 2.1 | 6.4 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 2.0 | 6.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 2.0 | 8.1 | GO:0070404 | NADH binding(GO:0070404) |
| 2.0 | 11.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.9 | 59.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.9 | 7.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 1.9 | 11.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 1.9 | 7.6 | GO:0016160 | amylase activity(GO:0016160) |
| 1.9 | 39.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 1.8 | 7.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 1.8 | 7.3 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) |
| 1.8 | 7.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 1.8 | 12.6 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 1.8 | 7.1 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 1.7 | 20.7 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 1.6 | 6.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 1.6 | 12.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.6 | 6.2 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 1.5 | 4.6 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 1.4 | 18.7 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 1.4 | 14.4 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 1.4 | 7.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 1.4 | 4.2 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 1.4 | 74.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 1.3 | 5.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 1.3 | 6.5 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 1.3 | 2.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 1.1 | 7.9 | GO:0031404 | chloride ion binding(GO:0031404) |
| 1.1 | 6.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.1 | 11.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.1 | 8.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 1.1 | 7.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 1.0 | 6.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.0 | 14.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 1.0 | 10.4 | GO:0019841 | retinol binding(GO:0019841) |
| 1.0 | 7.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 1.0 | 5.1 | GO:0016918 | retinal binding(GO:0016918) |
| 1.0 | 118.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 1.0 | 2.9 | GO:0016509 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 1.0 | 5.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 1.0 | 2.9 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.9 | 18.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.9 | 7.5 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.9 | 2.6 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.9 | 4.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.9 | 13.0 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.8 | 5.0 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.8 | 17.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.8 | 5.0 | GO:0070728 | leucine binding(GO:0070728) |
| 0.8 | 4.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.8 | 2.5 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.8 | 17.2 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.8 | 2.4 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.8 | 4.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.8 | 3.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.8 | 10.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.8 | 16.8 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.8 | 9.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.8 | 13.9 | GO:0001848 | complement binding(GO:0001848) |
| 0.7 | 2.2 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.7 | 4.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.7 | 1.5 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.7 | 8.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.7 | 11.4 | GO:0010181 | FMN binding(GO:0010181) |
| 0.7 | 2.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.7 | 2.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.7 | 2.8 | GO:0002046 | opsin binding(GO:0002046) |
| 0.7 | 2.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.7 | 12.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.7 | 5.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.7 | 10.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.6 | 5.8 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.6 | 3.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.6 | 1.8 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.6 | 15.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.6 | 1.7 | GO:0030977 | taurine binding(GO:0030977) |
| 0.5 | 1.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.5 | 15.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.5 | 11.2 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.5 | 1.0 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.5 | 3.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.5 | 1.5 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.5 | 5.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.5 | 7.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.5 | 1.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.5 | 1.8 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.5 | 9.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.5 | 7.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.4 | 30.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.4 | 1.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.4 | 5.7 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.4 | 2.6 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.4 | 11.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.4 | 18.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.4 | 1.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.4 | 0.8 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.4 | 2.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.4 | 2.4 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.4 | 1.2 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.4 | 2.3 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.4 | 1.9 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.4 | 1.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.4 | 5.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.4 | 0.7 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.4 | 5.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 1.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.4 | 12.0 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.4 | 1.8 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.4 | 3.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 5.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 1.0 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.3 | 6.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 7.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.3 | 6.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.3 | 3.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.3 | 3.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 3.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.3 | 10.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 2.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 1.5 | GO:0033041 | sweet taste receptor activity(GO:0033041) |
| 0.3 | 3.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.3 | 2.0 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.3 | 2.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 3.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 1.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.3 | 5.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.3 | 11.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.3 | 4.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 1.3 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.3 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 2.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.3 | 1.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 3.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 3.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 2.2 | GO:0008494 | translation activator activity(GO:0008494) translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.2 | 0.7 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.2 | 0.7 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 1.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 1.6 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.2 | 1.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 8.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.2 | 47.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 32.6 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.2 | 6.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 1.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 2.9 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 13.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 1.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 1.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 0.8 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.2 | 1.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 2.0 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.2 | 0.6 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.2 | 73.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 0.7 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 1.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 1.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 1.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.2 | 2.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 1.9 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 10.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.2 | 18.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 4.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 1.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 1.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 12.7 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 1.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 3.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.9 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 1.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 12.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 1.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.8 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 3.2 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 0.9 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.5 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 1.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 3.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 3.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 2.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 2.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 3.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 3.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 2.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 1.4 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.1 | 4.9 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 1.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 1.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 44.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 1.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.3 | GO:0016859 | cis-trans isomerase activity(GO:0016859) |
| 0.0 | 0.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 2.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.7 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 1.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.6 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 209.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.8 | 44.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.5 | 11.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.4 | 7.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.4 | 12.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 6.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.3 | 4.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 7.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 6.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 8.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 10.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 2.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 3.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 1.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.2 | 2.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 3.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 2.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 6.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 5.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 2.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 47.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 5.0 | 65.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 4.4 | 40.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 3.0 | 36.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 2.3 | 42.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 2.0 | 33.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.8 | 17.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 1.6 | 28.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.3 | 7.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.1 | 20.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 1.0 | 10.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 1.0 | 8.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.9 | 20.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.9 | 8.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.9 | 18.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.9 | 18.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.9 | 24.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.9 | 11.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.8 | 13.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.8 | 12.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.7 | 8.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.7 | 130.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.6 | 7.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.6 | 8.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.6 | 10.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.6 | 12.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.6 | 5.6 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.6 | 18.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.5 | 13.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.5 | 11.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.5 | 3.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.5 | 5.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.4 | 15.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.4 | 6.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 11.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.4 | 3.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 2.3 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.4 | 15.9 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.3 | 3.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 3.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 27.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 4.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.3 | 2.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 2.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.3 | 15.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 3.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.3 | 8.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 5.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 4.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 10.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 2.5 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.2 | 8.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 2.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 4.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 1.0 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 1.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 3.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 10.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 0.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 1.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 4.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.3 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 8.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 3.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 4.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.0 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.1 | 1.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 6.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 1.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.1 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.1 | 0.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 7.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |