Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Tal1
Z-value: 1.49
Transcription factors associated with Tal1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tal1
|
ENSRNOG00000025051 | TAL bHLH transcription factor 1, erythroid differentiation factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tal1 | rn6_v1_chr5_+_133864798_133864798 | 0.53 | 4.8e-25 | Click! |
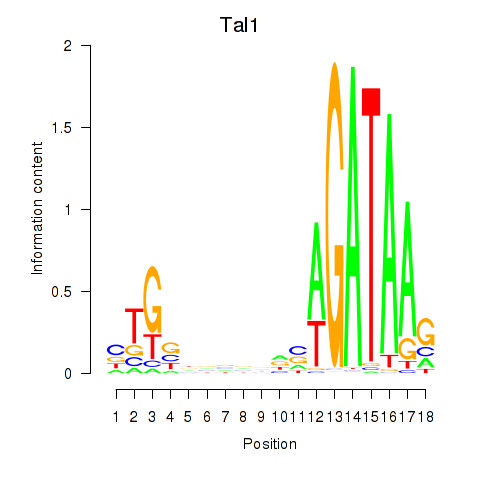
Activity profile of Tal1 motif
Sorted Z-values of Tal1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_151172206 | 89.06 |
ENSRNOT00000013778
|
Fgr
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr4_+_70614524 | 64.35 |
ENSRNOT00000041100
|
Prss3
|
protease, serine 3 |
| chr20_-_5806097 | 62.92 |
ENSRNOT00000000611
|
Clps
|
colipase |
| chr1_+_279798187 | 61.23 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr4_+_70689737 | 58.17 |
ENSRNOT00000018852
|
Prss2
|
protease, serine, 2 |
| chr9_-_23493081 | 56.81 |
ENSRNOT00000072144
|
Rhag
|
Rh-associated glycoprotein |
| chr1_+_279867034 | 53.34 |
ENSRNOT00000024164
|
Pnliprp1
|
pancreatic lipase-related protein 1 |
| chr20_-_2191640 | 52.58 |
ENSRNOT00000001016
|
Trim10
|
tripartite motif-containing 10 |
| chr10_-_90415070 | 49.75 |
ENSRNOT00000055179
|
Itga2b
|
integrin subunit alpha 2b |
| chr10_-_15590220 | 49.65 |
ENSRNOT00000048977
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr1_+_279896973 | 48.50 |
ENSRNOT00000068119
|
Pnliprp2
|
pancreatic lipase related protein 2 |
| chr4_+_57855416 | 47.12 |
ENSRNOT00000029608
|
Cpa2
|
carboxypeptidase A2 |
| chrX_-_23187341 | 47.08 |
ENSRNOT00000000180
|
Alas2
|
5'-aminolevulinate synthase 2 |
| chr7_+_121841855 | 43.01 |
ENSRNOT00000024673
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr4_+_117744075 | 40.27 |
ENSRNOT00000081173
|
Add2
|
adducin 2 |
| chr4_+_101645731 | 40.19 |
ENSRNOT00000087901
|
AABR07060953.1
|
|
| chr4_+_117743710 | 39.20 |
ENSRNOT00000021491
|
Add2
|
adducin 2 |
| chr4_-_103050006 | 38.95 |
ENSRNOT00000092130
|
LOC100911032
|
uncharacterized LOC100911032 |
| chr1_+_73040901 | 38.07 |
ENSRNOT00000071927
|
Gp6
|
glycoprotein 6 (platelet) |
| chr11_-_44049648 | 37.81 |
ENSRNOT00000002257
|
Cpox
|
coproporphyrinogen oxidase |
| chr4_-_70747226 | 36.71 |
ENSRNOT00000044960
|
LOC102554637
|
anionic trypsin-2-like |
| chr8_+_22047697 | 36.08 |
ENSRNOT00000067741
|
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr16_+_75572070 | 35.54 |
ENSRNOT00000043486
|
Defb52
|
defensin beta 52 |
| chr10_-_15603649 | 34.32 |
ENSRNOT00000051483
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr17_-_31706523 | 34.17 |
ENSRNOT00000071312
|
AABR07027447.1
|
|
| chr2_+_190073815 | 33.41 |
ENSRNOT00000015473
|
S100a8
|
S100 calcium binding protein A8 |
| chr14_+_22517774 | 33.36 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr2_+_188449210 | 31.84 |
ENSRNOT00000027700
|
Pklr
|
pyruvate kinase, liver and RBC |
| chr10_+_106785077 | 30.66 |
ENSRNOT00000075047
|
Tmc8
|
transmembrane channel-like 8 |
| chr15_-_28217614 | 29.10 |
ENSRNOT00000040857
|
Rnase3
|
ribonuclease A family member 3 |
| chr13_-_77896697 | 28.95 |
ENSRNOT00000003452
|
Tnn
|
tenascin N |
| chr2_-_173668555 | 27.75 |
ENSRNOT00000013452
|
Serpini2
|
serpin family I member 2 |
| chr3_+_17889972 | 26.14 |
ENSRNOT00000073021
|
AABR07051611.1
|
|
| chr13_+_83972212 | 25.97 |
ENSRNOT00000004394
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr17_-_31780120 | 25.67 |
ENSRNOT00000058388
|
AABR07027450.1
|
|
| chr15_-_28241198 | 25.09 |
ENSRNOT00000086535
|
LOC100361866
|
eosinophil cationic protein-like |
| chr10_+_70417108 | 24.66 |
ENSRNOT00000079325
|
Slfn4
|
schlafen 4 |
| chr16_-_19918644 | 24.41 |
ENSRNOT00000083345
ENSRNOT00000023926 |
Plvap
|
plasmalemma vesicle associated protein |
| chr10_+_70411738 | 24.12 |
ENSRNOT00000078046
|
Slfn4
|
schlafen 4 |
| chr11_+_85561460 | 23.58 |
ENSRNOT00000075455
|
AABR07072262.1
|
|
| chr3_-_48372583 | 22.95 |
ENSRNOT00000040482
ENSRNOT00000077788 ENSRNOT00000085426 |
Dpp4
|
dipeptidylpeptidase 4 |
| chr10_-_110232843 | 22.92 |
ENSRNOT00000054934
|
Cd7
|
Cd7 molecule |
| chr19_+_25815207 | 22.91 |
ENSRNOT00000003980
|
Lyl1
|
LYL1, basic helix-loop-helix family member |
| chr4_+_102351036 | 22.33 |
ENSRNOT00000079277
|
AABR07060994.1
|
|
| chr5_+_153197459 | 21.71 |
ENSRNOT00000023187
|
Rhd
|
Rh blood group, D antigen |
| chr4_-_103115522 | 21.34 |
ENSRNOT00000074206
|
AABR07061044.1
|
|
| chr4_-_102124609 | 20.94 |
ENSRNOT00000048263
|
AABR07060979.1
|
|
| chr5_+_133864798 | 20.77 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr4_-_103331319 | 19.61 |
ENSRNOT00000086338
|
AABR07061057.2
|
|
| chr11_+_85618714 | 19.48 |
ENSRNOT00000074614
|
AC109901.1
|
|
| chr1_-_98570949 | 19.42 |
ENSRNOT00000033648
|
Siglec5
|
sialic acid binding Ig-like lectin 5 |
| chr15_-_28204691 | 19.34 |
ENSRNOT00000048941
|
Rnase17
|
ribonuclease 17 |
| chr8_-_119326938 | 19.34 |
ENSRNOT00000044467
|
Ccrl2
|
C-C motif chemokine receptor like 2 |
| chr1_+_227640680 | 19.03 |
ENSRNOT00000033613
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C |
| chr7_-_101138860 | 18.40 |
ENSRNOT00000077137
|
AABR07058124.4
|
|
| chr3_+_119561290 | 18.37 |
ENSRNOT00000015843
|
Blvra
|
biliverdin reductase A |
| chr1_-_148119857 | 18.36 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr7_+_118685181 | 18.31 |
ENSRNOT00000068221
|
Apol3
|
apolipoprotein L, 3 |
| chr15_-_52399074 | 18.09 |
ENSRNOT00000018440
|
Xpo7
|
exportin 7 |
| chr20_+_9948908 | 18.00 |
ENSRNOT00000001541
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr15_+_57221292 | 17.88 |
ENSRNOT00000014502
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr14_+_44889287 | 17.79 |
ENSRNOT00000091312
ENSRNOT00000032273 |
Tmem156
|
transmembrane protein 156 |
| chr6_+_128050250 | 17.54 |
ENSRNOT00000077517
ENSRNOT00000013961 |
LOC500712
|
Ab1-233 |
| chr7_+_138707426 | 17.53 |
ENSRNOT00000037874
|
Pced1b
|
PC-esterase domain containing 1B |
| chr4_+_14070553 | 16.87 |
ENSRNOT00000077505
|
Cd36
|
CD36 molecule |
| chr1_+_189359853 | 16.83 |
ENSRNOT00000055083
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr15_+_28260411 | 16.51 |
ENSRNOT00000021645
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr8_-_22974321 | 16.51 |
ENSRNOT00000017369
|
Epor
|
erythropoietin receptor |
| chr7_+_118685022 | 16.37 |
ENSRNOT00000089135
|
Apol3
|
apolipoprotein L, 3 |
| chr7_-_115994783 | 16.30 |
ENSRNOT00000036268
|
Ly6d
|
lymphocyte antigen 6 complex, locus D |
| chr14_-_84189266 | 16.26 |
ENSRNOT00000005934
|
Tcn2
|
transcobalamin 2 |
| chr2_-_187909394 | 16.18 |
ENSRNOT00000032355
|
Rab25
|
RAB25, member RAS oncogene family |
| chrX_-_135250519 | 16.18 |
ENSRNOT00000044487
|
Elf4
|
E74 like ETS transcription factor 4 |
| chr8_-_33661049 | 15.77 |
ENSRNOT00000068037
|
Fli1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_+_48303366 | 15.49 |
ENSRNOT00000009973
|
Gabrr2
|
gamma-aminobutyric acid type A receptor rho 2 subunit |
| chr18_+_86299463 | 15.39 |
ENSRNOT00000058152
|
Cd226
|
CD226 molecule |
| chr4_-_165541314 | 15.29 |
ENSRNOT00000013833
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr4_+_102643934 | 15.22 |
ENSRNOT00000058389
|
LOC100911032
|
uncharacterized LOC100911032 |
| chr3_+_19366370 | 14.60 |
ENSRNOT00000086557
|
AABR07051689.1
|
|
| chr4_-_165195489 | 14.60 |
ENSRNOT00000079429
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr1_-_258877045 | 13.87 |
ENSRNOT00000071633
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr18_-_25314047 | 13.70 |
ENSRNOT00000079778
|
AABR07031674.4
|
|
| chr8_-_71728654 | 13.02 |
ENSRNOT00000031207
ENSRNOT00000091751 |
Snx22
|
sorting nexin 22 |
| chr6_-_142178771 | 12.90 |
ENSRNOT00000071577
|
Ighv12-3
|
immunoglobulin heavy variable V12-3 |
| chr12_-_45116665 | 12.55 |
ENSRNOT00000089043
|
Taok3
|
TAO kinase 3 |
| chr5_-_160423811 | 12.51 |
ENSRNOT00000018864
|
Efhd2
|
EF-hand domain family, member D2 |
| chr1_+_214454090 | 12.47 |
ENSRNOT00000049448
|
Tspan4
|
tetraspanin 4 |
| chr8_-_48672732 | 12.03 |
ENSRNOT00000079275
|
Hmbs
|
hydroxymethylbilane synthase |
| chr18_+_44716226 | 11.92 |
ENSRNOT00000086431
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr3_-_72171078 | 11.82 |
ENSRNOT00000009817
|
Serping1
|
serpin family G member 1 |
| chr13_-_83202864 | 11.79 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr1_+_81499821 | 11.74 |
ENSRNOT00000027100
|
Lypd3
|
Ly6/Plaur domain containing 3 |
| chr17_-_43821536 | 11.71 |
ENSRNOT00000072286
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr16_+_10267482 | 11.35 |
ENSRNOT00000085255
|
Gdf2
|
growth differentiation factor 2 |
| chr2_+_178679041 | 11.14 |
ENSRNOT00000013524
|
Fam198b
|
family with sequence similarity 198, member B |
| chr4_+_102290338 | 10.96 |
ENSRNOT00000011067
|
AABR07060992.1
|
|
| chr2_+_212257225 | 10.87 |
ENSRNOT00000077883
|
Vav3
|
vav guanine nucleotide exchange factor 3 |
| chrX_-_13279082 | 10.77 |
ENSRNOT00000051898
ENSRNOT00000060857 |
Tspan7
|
tetraspanin 7 |
| chr17_+_81352700 | 10.75 |
ENSRNOT00000024736
|
Mrc1
|
mannose receptor, C type 1 |
| chr1_+_282265370 | 10.61 |
ENSRNOT00000015687
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr16_-_10706073 | 10.54 |
ENSRNOT00000089114
|
Fam25a
|
family with sequence similarity 25, member A |
| chr1_+_72784966 | 10.49 |
ENSRNOT00000090065
ENSRNOT00000041527 |
Tmem86b
|
transmembrane protein 86B |
| chr4_-_103569159 | 10.29 |
ENSRNOT00000084036
|
AABR07061072.1
|
|
| chr18_-_74485139 | 10.15 |
ENSRNOT00000022598
|
Slc14a1
|
solute carrier family 14 member 1 |
| chr2_-_193330699 | 10.02 |
ENSRNOT00000012684
|
Crct1
|
cysteine-rich C-terminal 1 |
| chr11_-_80981415 | 9.74 |
ENSRNOT00000002499
ENSRNOT00000002496 |
St6gal1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr15_+_57290849 | 9.74 |
ENSRNOT00000014909
|
Cpb2
|
carboxypeptidase B2 |
| chr4_+_155088317 | 9.43 |
ENSRNOT00000088728
|
M6pr
|
mannose-6-phosphate receptor, cation dependent |
| chr5_-_76632462 | 9.35 |
ENSRNOT00000085109
|
Susd1
|
sushi domain containing 1 |
| chr15_+_34234755 | 9.29 |
ENSRNOT00000059987
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr9_+_98438439 | 9.29 |
ENSRNOT00000027220
|
Scly
|
selenocysteine lyase |
| chr1_+_260732485 | 9.27 |
ENSRNOT00000076842
ENSRNOT00000076073 |
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr15_-_28155291 | 9.17 |
ENSRNOT00000050820
|
Rnase2
|
ribonuclease A family member 2 |
| chr3_+_5519990 | 8.87 |
ENSRNOT00000070873
ENSRNOT00000007640 |
Adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr2_+_226563050 | 8.85 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr10_+_105630810 | 8.82 |
ENSRNOT00000064904
|
Prcd
|
photoreceptor disc component |
| chr11_+_85030872 | 8.79 |
ENSRNOT00000074815
|
LOC100911043
|
olfactory receptor 2L3-like |
| chr7_+_3216497 | 8.56 |
ENSRNOT00000008909
|
Mmp19
|
matrix metallopeptidase 19 |
| chr9_-_26707571 | 8.47 |
ENSRNOT00000080948
|
AABR07067023.1
|
|
| chr12_+_40244081 | 8.45 |
ENSRNOT00000030583
|
Sh2b3
|
SH2B adaptor protein 3 |
| chr14_-_44225713 | 8.22 |
ENSRNOT00000049161
|
LOC680579
|
similar to ribosomal protein L14 |
| chr2_+_178679395 | 7.93 |
ENSRNOT00000088686
|
Fam198b
|
family with sequence similarity 198, member B |
| chr3_-_148722710 | 7.73 |
ENSRNOT00000090919
ENSRNOT00000068592 |
Plagl2
|
PLAG1 like zinc finger 2 |
| chr11_+_31640407 | 7.66 |
ENSRNOT00000029985
ENSRNOT00000091472 |
Ifnar1
|
interferon alpha and beta receptor subunit 1 |
| chr7_-_116504853 | 7.51 |
ENSRNOT00000056557
|
RGD1565410
|
similar to Ly6-C antigen gene |
| chr7_-_11444786 | 7.47 |
ENSRNOT00000027323
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr8_+_64088913 | 7.26 |
ENSRNOT00000013022
|
Adpgk
|
ADP-dependent glucokinase |
| chr19_-_838099 | 7.12 |
ENSRNOT00000014234
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr1_-_221370322 | 7.07 |
ENSRNOT00000028431
|
Capn1
|
calpain 1 |
| chr8_+_96564877 | 6.87 |
ENSRNOT00000017879
|
Mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chrX_-_135342996 | 6.83 |
ENSRNOT00000084848
ENSRNOT00000008503 |
Aifm1
|
apoptosis inducing factor, mitochondria associated 1 |
| chr3_-_123702732 | 6.56 |
ENSRNOT00000028859
|
RGD1311739
|
similar to RIKEN cDNA 1700037H04 |
| chr10_-_34961608 | 6.55 |
ENSRNOT00000033056
|
LOC103689931
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr5_-_115222075 | 6.53 |
ENSRNOT00000058163
ENSRNOT00000012503 |
LOC100912642
|
cytochrome P450 2J3-like |
| chr1_+_165482155 | 6.49 |
ENSRNOT00000024005
|
Ucp3
|
uncoupling protein 3 |
| chr1_-_80178708 | 6.45 |
ENSRNOT00000088676
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr14_+_43810119 | 6.38 |
ENSRNOT00000003327
|
Rbm47
|
RNA binding motif protein 47 |
| chr13_-_36290531 | 6.38 |
ENSRNOT00000071388
|
Steap3
|
STEAP3 metalloreductase |
| chr3_-_123236535 | 6.17 |
ENSRNOT00000028839
|
Slc4a11
|
solute carrier family 4 member 11 |
| chr1_+_214016897 | 6.13 |
ENSRNOT00000075588
|
LOC100911766
|
tetraspanin-4-like |
| chr19_-_43528851 | 6.10 |
ENSRNOT00000036972
|
Mlkl
|
mixed lineage kinase domain like pseudokinase |
| chr12_+_48403797 | 6.01 |
ENSRNOT00000044751
|
Ssh1
|
slingshot protein phosphatase 1 |
| chr20_-_2211995 | 5.99 |
ENSRNOT00000077201
|
Trim26
|
tripartite motif-containing 26 |
| chr4_+_147333056 | 5.97 |
ENSRNOT00000012137
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
| chr10_+_57260680 | 5.86 |
ENSRNOT00000035445
|
Gp1ba
|
glycoprotein Ib platelet alpha subunit |
| chr20_-_5056474 | 5.74 |
ENSRNOT00000076375
ENSRNOT00000037191 ENSRNOT00000076863 |
G6b
|
immunoreceptor tyrosine-based inhibitory motif (ITIM) containing platelet receptor |
| chr8_+_42347811 | 5.64 |
ENSRNOT00000073897
|
LOC100912186
|
olfactory receptor 8B3-like |
| chr16_-_49820235 | 5.46 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr3_+_125320680 | 5.46 |
ENSRNOT00000028891
|
LOC681292
|
hypothetical protein LOC681292 |
| chr10_-_89374516 | 5.43 |
ENSRNOT00000028084
|
Vat1
|
vesicle amine transport 1 |
| chr4_+_154215250 | 5.43 |
ENSRNOT00000072465
|
Mug2
|
murinoglobulin 2 |
| chr14_+_107349414 | 5.40 |
ENSRNOT00000012255
|
LOC690096
|
similar to ribosomal protein L28 |
| chr14_-_110883784 | 5.39 |
ENSRNOT00000086299
ENSRNOT00000068190 |
Vrk2
|
vaccinia related kinase 2 |
| chr7_-_115981731 | 5.33 |
ENSRNOT00000036549
|
RGD1308195
|
similar to secreted Ly6/uPAR related protein 2 |
| chr16_+_2634603 | 5.25 |
ENSRNOT00000019113
|
Hesx1
|
HESX homeobox 1 |
| chr6_+_58388333 | 5.25 |
ENSRNOT00000036337
|
LOC680646
|
similar to 40S ribosomal protein S2 |
| chr16_-_68968248 | 5.16 |
ENSRNOT00000016885
|
Eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr9_+_81535483 | 5.09 |
ENSRNOT00000077285
|
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr10_+_34938667 | 5.05 |
ENSRNOT00000086443
ENSRNOT00000059104 |
LOC103690164
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr19_+_55929609 | 5.02 |
ENSRNOT00000068231
ENSRNOT00000077701 |
Cpne7
|
copine 7 |
| chr10_-_37055282 | 4.89 |
ENSRNOT00000086193
ENSRNOT00000065584 |
Hnrnpab
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr1_-_37990007 | 4.48 |
ENSRNOT00000081054
|
AABR07001100.1
|
|
| chr8_-_66863476 | 4.38 |
ENSRNOT00000018820
|
Rplp1
|
ribosomal protein lateral stalk subunit P1 |
| chr7_-_5195955 | 4.36 |
ENSRNOT00000040141
|
Olr896
|
olfactory receptor 896 |
| chr18_+_56379890 | 4.33 |
ENSRNOT00000078764
|
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr6_+_27975849 | 4.21 |
ENSRNOT00000060810
|
Dtnb
|
dystrobrevin, beta |
| chr16_+_31734944 | 4.14 |
ENSRNOT00000059673
|
Palld
|
palladin, cytoskeletal associated protein |
| chr5_-_62001196 | 4.08 |
ENSRNOT00000012602
|
Trmo
|
tRNA methyltransferase O |
| chr5_+_164808323 | 4.08 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr17_+_39156840 | 4.07 |
ENSRNOT00000022447
|
Prl7b1
|
prolactin family 7, subfamily b, member 1 |
| chr10_-_110182291 | 4.02 |
ENSRNOT00000015178
ENSRNOT00000054936 |
Csnk1d
|
casein kinase 1, delta |
| chr3_-_73755510 | 3.85 |
ENSRNOT00000083628
|
Olr502
|
olfactory receptor 502 |
| chr13_-_45068077 | 3.85 |
ENSRNOT00000004969
|
Mcm6
|
minichromosome maintenance complex component 6 |
| chr10_+_31813814 | 3.68 |
ENSRNOT00000009573
|
Havcr1
|
hepatitis A virus cellular receptor 1 |
| chrX_-_16532089 | 3.66 |
ENSRNOT00000093156
|
Dgkk
|
diacylglycerol kinase kappa |
| chr15_-_45524582 | 3.65 |
ENSRNOT00000081912
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr3_-_73510015 | 3.63 |
ENSRNOT00000072248
|
Olr483
|
olfactory receptor 483 |
| chr1_-_72941869 | 3.59 |
ENSRNOT00000032306
|
Eps8l1
|
EPS8-like 1 |
| chr8_-_116856548 | 3.57 |
ENSRNOT00000043825
|
Rnf123
|
ring finger protein 123 |
| chr10_+_37032043 | 3.56 |
ENSRNOT00000037303
ENSRNOT00000091242 |
Phykpl
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr4_+_140377565 | 3.54 |
ENSRNOT00000082723
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr10_-_87407634 | 3.48 |
ENSRNOT00000016657
|
Krt23
|
keratin 23 |
| chr9_+_41006800 | 3.43 |
ENSRNOT00000079413
ENSRNOT00000034214 |
Prss39
|
protease, serine, 39 |
| chr16_+_61926586 | 3.39 |
ENSRNOT00000058884
|
AABR07026059.1
|
|
| chr10_+_59613398 | 3.35 |
ENSRNOT00000025553
|
Ncbp3
|
nuclear cap binding subunit 3 |
| chr1_+_171797516 | 3.34 |
ENSRNOT00000088110
|
Ppfibp2
|
PPFIA binding protein 2 |
| chr1_-_175895510 | 3.27 |
ENSRNOT00000064535
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr2_+_188748359 | 3.20 |
ENSRNOT00000028038
|
Shc1
|
SHC adaptor protein 1 |
| chr10_+_87808493 | 3.12 |
ENSRNOT00000065405
|
LOC680428
|
hypothetical protein LOC680428 |
| chr6_+_102477036 | 3.10 |
ENSRNOT00000087132
|
Rad51b
|
RAD51 paralog B |
| chr14_+_11988671 | 3.04 |
ENSRNOT00000003207
|
Rasgef1b
|
RasGEF domain family, member 1B |
| chr3_+_11207542 | 3.02 |
ENSRNOT00000013546
|
Prrc2b
|
proline-rich coiled-coil 2B |
| chr18_+_16544508 | 2.91 |
ENSRNOT00000020601
|
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr3_-_76279104 | 2.86 |
ENSRNOT00000007796
|
Olr608
|
olfactory receptor 608 |
| chr5_-_137014375 | 2.83 |
ENSRNOT00000066165
ENSRNOT00000091866 |
Kdm4a
|
lysine demethylase 4A |
| chr3_+_76179214 | 2.79 |
ENSRNOT00000071281
|
LOC686677
|
similar to olfactory receptor 1161 |
| chr8_-_63291966 | 2.78 |
ENSRNOT00000077762
|
AABR07070278.1
|
|
| chr10_-_88107232 | 2.72 |
ENSRNOT00000019330
ENSRNOT00000076770 |
Krt9
|
keratin 9 |
| chr3_-_12467662 | 2.68 |
ENSRNOT00000051893
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr10_-_16788126 | 2.67 |
ENSRNOT00000077386
|
Atp6v0e1
|
ATPase H+ transporting V0 subunit e1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tal1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.8 | 47.1 | GO:1901423 | response to benzene(GO:1901423) |
| 8.1 | 56.8 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 7.6 | 22.9 | GO:0061744 | motor behavior(GO:0061744) |
| 7.2 | 29.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 6.9 | 48.5 | GO:0019374 | galactolipid metabolic process(GO:0019374) |
| 6.9 | 20.8 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 5.6 | 61.2 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 5.4 | 80.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 5.1 | 15.4 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 5.0 | 49.8 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 4.5 | 89.1 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 4.1 | 16.5 | GO:1904373 | response to kainic acid(GO:1904373) |
| 3.9 | 11.8 | GO:0071663 | immunoglobulin production in mucosal tissue(GO:0002426) T-helper 1 cell activation(GO:0035711) granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) regulation of thymocyte migration(GO:2000410) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 3.7 | 18.4 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 3.3 | 84.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 3.2 | 9.7 | GO:1990743 | protein sialylation(GO:1990743) |
| 3.2 | 9.7 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 3.2 | 31.8 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 3.1 | 56.1 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 2.8 | 14.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 2.8 | 16.9 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 2.8 | 58.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 2.7 | 16.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 2.5 | 22.9 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 2.4 | 58.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 2.3 | 6.9 | GO:0042560 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 2.1 | 6.4 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 2.0 | 6.0 | GO:2000230 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 2.0 | 33.4 | GO:0018119 | leukocyte migration involved in inflammatory response(GO:0002523) protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 1.8 | 29.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 1.8 | 30.7 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 1.7 | 10.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 1.6 | 6.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.6 | 21.7 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 1.5 | 17.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 1.4 | 24.4 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 1.4 | 6.8 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 1.4 | 4.1 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 1.3 | 5.4 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 1.3 | 9.3 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 1.3 | 10.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 1.3 | 16.3 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 1.2 | 4.9 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 1.2 | 6.0 | GO:1904717 | positive regulation of receptor clustering(GO:1903911) regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 1.2 | 32.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.1 | 4.3 | GO:0072276 | cell migration involved in vasculogenesis(GO:0035441) VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 1.0 | 9.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 1.0 | 5.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 1.0 | 11.8 | GO:2000258 | complement activation, lectin pathway(GO:0001867) negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.9 | 15.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.9 | 55.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.9 | 5.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.9 | 7.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.9 | 4.4 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 0.9 | 10.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.9 | 60.3 | GO:0032094 | response to food(GO:0032094) |
| 0.9 | 8.6 | GO:0001554 | luteolysis(GO:0001554) |
| 0.8 | 16.2 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.8 | 11.4 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.8 | 5.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.7 | 2.2 | GO:0042335 | cuticle development(GO:0042335) |
| 0.7 | 3.7 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.7 | 3.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.7 | 15.3 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.7 | 11.7 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.7 | 48.8 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.6 | 7.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.6 | 3.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.6 | 19.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.6 | 7.7 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.6 | 1.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.6 | 22.9 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.5 | 64.3 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.5 | 47.0 | GO:0031016 | pancreas development(GO:0031016) |
| 0.5 | 10.9 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.5 | 2.1 | GO:0002921 | negative regulation of humoral immune response(GO:0002921) |
| 0.5 | 3.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.5 | 12.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.5 | 1.4 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.5 | 70.1 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.5 | 8.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.5 | 6.5 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.4 | 15.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.4 | 7.7 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.4 | 4.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.4 | 0.4 | GO:1901671 | positive regulation of superoxide dismutase activity(GO:1901671) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.4 | 19.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.4 | 8.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.4 | 11.9 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.3 | 4.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.3 | 2.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.3 | 34.7 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.3 | 18.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.3 | 2.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 14.6 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.2 | 0.5 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.2 | 2.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 6.1 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.2 | 16.8 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.2 | 6.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 3.1 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.2 | 4.0 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.2 | 2.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 1.0 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 5.3 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.2 | 1.1 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.1 | 0.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 7.5 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 3.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.6 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 6.5 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.1 | 5.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 1.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 3.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 2.7 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 1.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 1.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.3 | GO:0061526 | acetylcholine secretion(GO:0061526) |
| 0.1 | 3.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 1.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 3.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.9 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0002155 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.1 | GO:1904823 | purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 1.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 3.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.0 | GO:0046487 | glyoxylate metabolic process(GO:0046487) negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 1.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.4 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.0 | 84.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 7.9 | 79.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 4.8 | 38.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 2.6 | 48.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 2.1 | 14.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 1.8 | 87.2 | GO:0016235 | aggresome(GO:0016235) |
| 1.5 | 49.7 | GO:0008305 | integrin complex(GO:0008305) |
| 1.2 | 16.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.9 | 9.8 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.9 | 3.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.8 | 5.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.8 | 3.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.7 | 15.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.7 | 3.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.6 | 17.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.6 | 3.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.6 | 4.9 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.6 | 44.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.4 | 11.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.4 | 5.9 | GO:0031362 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 3.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 9.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.3 | 2.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 4.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.3 | 9.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.3 | 507.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.3 | 6.5 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.3 | 20.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.3 | 18.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.3 | 24.4 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 6.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 2.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 2.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 31.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 16.2 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 1.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 3.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 4.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 4.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 36.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 2.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 8.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.4 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 11.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 6.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 6.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 14.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 4.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 21.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 7.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 3.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 9.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 35.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 9.6 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 3.4 | GO:0016604 | nuclear body(GO:0016604) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 28.0 | 84.0 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 15.7 | 47.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 14.8 | 89.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 8.3 | 49.7 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 8.0 | 160.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 8.0 | 31.8 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 7.1 | 56.8 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 5.6 | 33.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 5.4 | 37.8 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 4.0 | 12.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 3.8 | 38.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 3.4 | 16.9 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 2.6 | 10.3 | GO:0015265 | urea channel activity(GO:0015265) |
| 2.6 | 7.7 | GO:0019961 | interferon binding(GO:0019961) |
| 2.4 | 9.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 2.3 | 22.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 2.3 | 6.9 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 2.3 | 18.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 2.1 | 16.8 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 2.0 | 54.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 1.8 | 14.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 1.8 | 10.6 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 1.6 | 6.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.6 | 79.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 1.6 | 6.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 1.5 | 10.5 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 1.4 | 4.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.1 | 15.5 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 1.1 | 32.2 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.0 | 4.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 1.0 | 20.2 | GO:0005537 | mannose binding(GO:0005537) |
| 1.0 | 9.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.9 | 3.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.9 | 99.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.9 | 6.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.8 | 5.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.8 | 9.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.8 | 4.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.8 | 31.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.8 | 3.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.8 | 92.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.8 | 21.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.7 | 3.6 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.7 | 18.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.7 | 14.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.7 | 20.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.6 | 16.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.6 | 11.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.6 | 8.4 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.6 | 122.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.4 | 5.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 2.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 11.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.4 | 19.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.4 | 6.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 2.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.4 | 1.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 5.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 4.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 1.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 8.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.3 | 6.2 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.3 | 2.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.3 | 7.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 15.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 19.4 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.3 | 29.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 5.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 3.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 3.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 11.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 3.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.2 | 7.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 1.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 7.7 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.2 | 3.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 15.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.2 | 12.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.2 | 0.8 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 17.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 3.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 4.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 3.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 2.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 1.4 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.1 | 6.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.3 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 2.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 4.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 38.7 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.1 | 11.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 23.9 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.1 | 6.0 | GO:1990837 | sequence-specific double-stranded DNA binding(GO:1990837) |
| 0.1 | 32.8 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 5.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 2.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 4.1 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 6.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 7.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 10.0 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 43.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 3.2 | 138.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 1.6 | 41.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.4 | 33.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.8 | 184.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.8 | 7.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.7 | 14.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.7 | 25.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.6 | 31.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.6 | 2.9 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.5 | 12.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.4 | 11.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.4 | 4.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.4 | 6.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.4 | 10.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.4 | 22.9 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.3 | 10.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.3 | 4.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.3 | 6.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 3.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.2 | 6.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 4.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 6.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 5.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 9.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 5.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 1.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 115.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 6.5 | 58.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 4.2 | 89.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 3.5 | 172.7 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 2.8 | 49.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 1.9 | 49.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 1.8 | 66.9 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 1.4 | 20.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 1.3 | 31.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 1.1 | 51.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.9 | 17.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.6 | 9.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.6 | 15.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.5 | 8.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.5 | 7.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.4 | 9.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 10.9 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.4 | 27.9 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.4 | 3.2 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.3 | 3.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 9.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 11.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 4.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 10.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 2.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 3.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 5.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 3.2 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.1 | 0.8 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 4.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 4.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |