Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
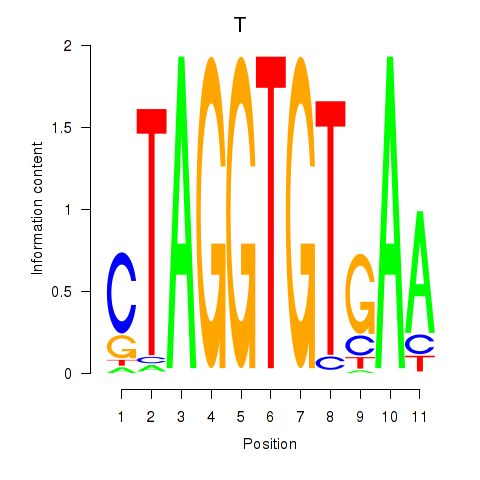
Results for T
Z-value: 1.20
Transcription factors associated with T
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
T
|
ENSRNOG00000012229 | T brachyury transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| T | rn6_v1_chr1_-_52894832_52894832 | -0.34 | 7.7e-10 | Click! |
Activity profile of T motif
Sorted Z-values of T motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_23289266 | 88.90 |
ENSRNOT00000061708
|
Bhmt2
|
betaine-homocysteine S-methyltransferase 2 |
| chr1_+_238222521 | 74.90 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr20_-_45126062 | 60.65 |
ENSRNOT00000000720
|
RGD1310495
|
similar to KIAA1919 protein |
| chr11_+_74057361 | 48.78 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chr1_-_240601744 | 48.44 |
ENSRNOT00000024093
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr2_+_68821004 | 47.81 |
ENSRNOT00000083713
|
Egf
|
epidermal growth factor |
| chr1_+_222310920 | 47.43 |
ENSRNOT00000091465
|
Macrod1
|
MACRO domain containing 1 |
| chrX_+_110818716 | 42.12 |
ENSRNOT00000086308
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_148240504 | 42.09 |
ENSRNOT00000085373
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr6_-_127508452 | 38.92 |
ENSRNOT00000073709
|
LOC100909524
|
protein Z-dependent protease inhibitor-like |
| chr1_-_102780381 | 37.42 |
ENSRNOT00000080132
|
Saa4
|
serum amyloid A4 |
| chr15_+_10120206 | 35.33 |
ENSRNOT00000033048
|
Rarb
|
retinoic acid receptor, beta |
| chr4_-_148845267 | 31.34 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr2_-_210749991 | 27.92 |
ENSRNOT00000051261
ENSRNOT00000052403 |
Gstm6l
|
glutathione S-transferase, mu 6-like |
| chr12_+_40466495 | 27.82 |
ENSRNOT00000001816
|
Aldh2
|
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr16_-_81834945 | 27.24 |
ENSRNOT00000037806
|
F7
|
coagulation factor VII |
| chr14_+_22251499 | 26.60 |
ENSRNOT00000087991
ENSRNOT00000002705 |
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr2_-_188596222 | 25.29 |
ENSRNOT00000027920
|
Efna1
|
ephrin A1 |
| chr8_-_58195884 | 25.15 |
ENSRNOT00000010573
|
Acat1
|
acetyl-CoA acetyltransferase 1 |
| chr4_+_122365093 | 24.73 |
ENSRNOT00000024011
|
Klf15
|
Kruppel-like factor 15 |
| chr3_-_176706896 | 23.96 |
ENSRNOT00000017337
|
Ptk6
|
protein tyrosine kinase 6 |
| chr3_+_142383278 | 22.51 |
ENSRNOT00000017742
|
Foxa2
|
forkhead box A2 |
| chrX_-_111102464 | 22.11 |
ENSRNOT00000084176
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr9_-_63637677 | 20.34 |
ENSRNOT00000049259
|
Satb2
|
SATB homeobox 2 |
| chr3_+_61620192 | 19.49 |
ENSRNOT00000065426
|
Hoxd9
|
homeo box D9 |
| chr7_+_144577465 | 18.78 |
ENSRNOT00000021647
|
Hoxc10
|
homeo box C10 |
| chr2_+_127845034 | 16.85 |
ENSRNOT00000044804
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr3_-_172566010 | 16.35 |
ENSRNOT00000071913
|
Atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr8_+_72405748 | 16.31 |
ENSRNOT00000023952
|
Car12
|
carbonic anhydrase 12 |
| chr13_-_51297621 | 16.17 |
ENSRNOT00000030926
|
Ndufv3
|
NADH:ubiquinone oxidoreductase subunit V3 |
| chr13_+_49092306 | 15.96 |
ENSRNOT00000000039
|
Nuak2
|
NUAK family kinase 2 |
| chr10_-_61772250 | 15.89 |
ENSRNOT00000092338
ENSRNOT00000085394 ENSRNOT00000046110 |
Srr
|
serine racemase |
| chr13_-_25262469 | 15.73 |
ENSRNOT00000019921
|
Rnf152
|
ring finger protein 152 |
| chr16_+_20962227 | 15.39 |
ENSRNOT00000027615
|
Slc25a42
|
solute carrier family 25, member 42 |
| chr11_+_88699222 | 14.80 |
ENSRNOT00000084177
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chrX_-_111179152 | 14.78 |
ENSRNOT00000089115
|
Morc4
|
MORC family CW-type zinc finger 4 |
| chr1_+_256382791 | 14.77 |
ENSRNOT00000022549
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr10_-_82116621 | 13.05 |
ENSRNOT00000003977
ENSRNOT00000051497 ENSRNOT00000085451 |
Abcc3
|
ATP binding cassette subfamily C member 3 |
| chrX_-_75566481 | 12.98 |
ENSRNOT00000003714
|
Zdhhc15
|
zinc finger, DHHC-type containing 15 |
| chr2_-_210766501 | 12.02 |
ENSRNOT00000025800
|
Gstm3
|
glutathione S-transferase mu 3 |
| chr12_-_30314519 | 11.80 |
ENSRNOT00000001220
|
Zbed5
|
zinc finger, BED-type containing 5 |
| chr5_-_56536772 | 11.29 |
ENSRNOT00000060765
|
Ddx58
|
DEXD/H-box helicase 58 |
| chr14_-_87701884 | 10.49 |
ENSRNOT00000079338
|
Mospd1
|
motile sperm domain containing 1 |
| chr13_-_50509916 | 10.39 |
ENSRNOT00000076747
|
Ren
|
renin |
| chr18_-_3662654 | 9.90 |
ENSRNOT00000092854
ENSRNOT00000016167 |
Npc1
|
NPC intracellular cholesterol transporter 1 |
| chr1_-_213635546 | 9.68 |
ENSRNOT00000018861
|
Sirt3
|
sirtuin 3 |
| chr16_+_46731403 | 9.27 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chrX_+_54266687 | 8.74 |
ENSRNOT00000058309
|
LOC100363193
|
LRRGT00076-like |
| chr18_+_16616937 | 8.68 |
ENSRNOT00000093641
|
Mocos
|
molybdenum cofactor sulfurase |
| chr1_+_172264473 | 8.64 |
ENSRNOT00000050651
|
Olr246
|
olfactory receptor 246 |
| chr1_-_89559960 | 8.63 |
ENSRNOT00000092133
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr11_+_27208564 | 8.34 |
ENSRNOT00000002158
|
Map3k7cl
|
MAP3K7 C-terminal like |
| chr2_+_127459012 | 8.16 |
ENSRNOT00000093685
ENSRNOT00000072675 |
Intu
|
inturned planar cell polarity protein |
| chr1_+_197839430 | 8.13 |
ENSRNOT00000025043
|
Rabep2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr14_-_82975263 | 7.98 |
ENSRNOT00000024165
|
Slc5a1
|
solute carrier family 5 member 1 |
| chr15_+_34552410 | 7.63 |
ENSRNOT00000027802
|
Khnyn
|
KH and NYN domain containing |
| chr10_-_47630799 | 7.48 |
ENSRNOT00000057907
|
Slc47a2
|
solute carrier family 47 member 2 |
| chr16_-_49574314 | 7.34 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr15_+_107486555 | 7.15 |
ENSRNOT00000074277
|
AABR07019512.1
|
|
| chr3_+_151126591 | 7.01 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr20_+_47494424 | 6.90 |
ENSRNOT00000087718
|
Sec63
|
SEC63 homolog, protein translocation regulator |
| chr1_-_91832058 | 6.80 |
ENSRNOT00000079088
|
Nudt19
|
nudix hydrolase 19 |
| chr3_-_164728338 | 6.45 |
ENSRNOT00000035463
|
Fam65c
|
family with sequence similarity 65, member C |
| chr7_-_143187594 | 6.22 |
ENSRNOT00000011732
ENSRNOT00000083466 |
Krt84
|
keratin 84 |
| chr2_-_153140894 | 6.03 |
ENSRNOT00000075166
|
AABR07010840.1
|
|
| chr6_-_103605256 | 5.94 |
ENSRNOT00000006005
|
Dcaf5
|
DDB1 and CUL4 associated factor 5 |
| chr3_-_77792014 | 5.61 |
ENSRNOT00000091085
|
Olr671
|
olfactory receptor 671 |
| chr1_-_22404002 | 5.56 |
ENSRNOT00000044098
|
Taar8a
|
trace amine-associated receptor 8a |
| chr1_+_220243473 | 5.42 |
ENSRNOT00000084650
ENSRNOT00000027091 |
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr2_-_220663305 | 5.29 |
ENSRNOT00000090294
|
AABR07013057.1
|
|
| chr1_+_213636093 | 5.17 |
ENSRNOT00000019642
|
Psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr19_+_52374298 | 5.16 |
ENSRNOT00000092131
|
Atp2c2
|
ATPase secretory pathway Ca2+ transporting 2 |
| chr12_+_46869836 | 5.05 |
ENSRNOT00000084421
|
Sirt4
|
sirtuin 4 |
| chr13_+_56513286 | 5.00 |
ENSRNOT00000015596
|
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr8_-_36842370 | 4.90 |
ENSRNOT00000074011
|
Pate3
|
prostate and testis expressed 3 |
| chr19_+_39126589 | 4.84 |
ENSRNOT00000027561
|
Sntb2
|
syntrophin, beta 2 |
| chr8_-_22336794 | 4.79 |
ENSRNOT00000066340
|
Ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr3_-_118427851 | 4.78 |
ENSRNOT00000055995
|
Fam227b
|
family with sequence similarity 227, member B |
| chr3_-_149563476 | 4.77 |
ENSRNOT00000071869
|
AABR07054352.2
|
|
| chr1_+_172813394 | 4.68 |
ENSRNOT00000090059
|
Olr272
|
olfactory receptor 272 |
| chr10_-_13175863 | 4.61 |
ENSRNOT00000034380
|
Prss33
|
protease, serine, 33 |
| chr3_-_61636038 | 4.61 |
ENSRNOT00000086887
|
AABR07052559.1
|
|
| chr20_+_47494270 | 4.54 |
ENSRNOT00000036365
|
Sec63
|
SEC63 homolog, protein translocation regulator |
| chr12_-_35318685 | 4.43 |
ENSRNOT00000074199
|
AABR07036167.1
|
|
| chr7_+_6096690 | 4.42 |
ENSRNOT00000051694
|
Olr1006
|
olfactory receptor 1006 |
| chr7_+_78732904 | 4.42 |
ENSRNOT00000088433
|
LOC103690326
|
dendritic cell-specific transmembrane protein |
| chr6_-_21112734 | 4.39 |
ENSRNOT00000079819
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chr19_-_37907714 | 4.38 |
ENSRNOT00000026361
|
Ctrl
|
chymotrypsin-like |
| chr2_+_160291450 | 4.32 |
ENSRNOT00000075563
|
AABR07011278.1
|
|
| chr20_+_6356423 | 4.23 |
ENSRNOT00000000628
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr10_-_51778939 | 4.19 |
ENSRNOT00000078675
ENSRNOT00000057562 |
Myocd
|
myocardin |
| chr7_+_38945836 | 4.12 |
ENSRNOT00000006455
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr1_+_218487824 | 4.05 |
ENSRNOT00000017958
|
Mrgprd
|
MAS related GPR family member D |
| chr3_+_100768637 | 3.92 |
ENSRNOT00000083542
|
Bdnf
|
brain-derived neurotrophic factor |
| chr1_-_73121085 | 3.78 |
ENSRNOT00000073464
|
LOC684545
|
similar to NACHT, leucine rich repeat and PYD containing 2 |
| chr10_-_61045355 | 3.73 |
ENSRNOT00000048167
|
Olr1514
|
olfactory receptor 1514 |
| chr3_+_11607225 | 3.68 |
ENSRNOT00000072685
|
St6galnac4
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr5_+_148320438 | 3.56 |
ENSRNOT00000018742
|
Pef1
|
penta-EF hand domain containing 1 |
| chr3_+_160695911 | 3.54 |
ENSRNOT00000018510
|
Svs6
|
seminal vesicle secretory protein 6 |
| chr1_-_22596475 | 3.43 |
ENSRNOT00000021510
|
Taar1
|
trace-amine-associated receptor 1 |
| chr1_-_87937516 | 3.38 |
ENSRNOT00000087522
|
Eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr8_-_22173797 | 3.35 |
ENSRNOT00000042314
ENSRNOT00000079949 |
Cdc37
|
cell division cycle 37 |
| chr20_-_5931417 | 3.29 |
ENSRNOT00000092383
|
Slc26a8
|
solute carrier family 26 member 8 |
| chr2_-_1511591 | 3.29 |
ENSRNOT00000087906
ENSRNOT00000014709 |
Cast
|
calpastatin |
| chr3_+_113423693 | 3.28 |
ENSRNOT00000021091
|
Hypk
|
Huntingtin interacting protein K |
| chr2_+_211880262 | 3.22 |
ENSRNOT00000038315
|
Slc25a54
|
solute carrier family 25, member 54 |
| chr16_-_82999608 | 3.20 |
ENSRNOT00000078867
|
Tex29
|
testis expressed 29 |
| chr2_-_149754776 | 3.11 |
ENSRNOT00000035057
|
RGD1559622
|
similar to hypothetical protein C130079G13 |
| chr4_+_1584083 | 3.05 |
ENSRNOT00000087099
|
Olr1244
|
olfactory receptor 1244 |
| chr1_+_79181855 | 3.05 |
ENSRNOT00000037132
|
Psgb1
|
pregnancy-specific beta 1-glycoprotein |
| chr4_+_174181644 | 2.97 |
ENSRNOT00000011555
|
Capza3
|
capping actin protein of muscle Z-line alpha subunit 3 |
| chr8_-_124399494 | 2.88 |
ENSRNOT00000037883
|
Tgfbr2
|
transforming growth factor, beta receptor 2 |
| chr5_+_162323373 | 2.82 |
ENSRNOT00000033150
|
Aadacl3
|
arylacetamide deacetylase-like 3 |
| chr11_-_64968437 | 2.69 |
ENSRNOT00000059541
|
Cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr4_+_117679342 | 2.69 |
ENSRNOT00000021272
|
Figla
|
folliculogenesis specific bHLH transcription factor |
| chr11_+_81769178 | 2.64 |
ENSRNOT00000038826
|
Tbccd1
|
TBCC domain containing 1 |
| chr1_-_154165524 | 2.60 |
ENSRNOT00000023468
|
Hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chr18_+_38847632 | 2.59 |
ENSRNOT00000014916
|
LOC102554034
|
ELMO domain-containing protein 2-like |
| chr10_-_61088827 | 2.57 |
ENSRNOT00000047421
|
Olr1517
|
olfactory receptor 1517 |
| chr19_+_9622611 | 2.53 |
ENSRNOT00000061498
|
Slc38a7
|
solute carrier family 38, member 7 |
| chr16_-_82999827 | 2.51 |
ENSRNOT00000041223
|
Tex29
|
testis expressed 29 |
| chr1_+_168844875 | 2.44 |
ENSRNOT00000051398
|
AC107531.1
|
|
| chr9_-_99365192 | 2.37 |
ENSRNOT00000047012
|
RGD1564730
|
similar to ribosomal protein L36 |
| chr17_+_3924473 | 2.36 |
ENSRNOT00000061470
|
MGC114246
|
similar to cathepsin R |
| chr4_+_45120576 | 2.33 |
ENSRNOT00000077961
|
ST7
|
suppression of tumorigenicity 7 |
| chr18_-_60835739 | 2.33 |
ENSRNOT00000039569
|
LOC291543
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr7_+_129595192 | 2.31 |
ENSRNOT00000071151
|
Zdhhc25
|
zinc finger, DHHC-type containing 25 |
| chr1_-_215858034 | 2.26 |
ENSRNOT00000027656
|
Ins2
|
insulin 2 |
| chr7_+_77066955 | 2.15 |
ENSRNOT00000008700
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr2_+_205617220 | 2.15 |
ENSRNOT00000025365
|
Dennd2c
|
DENN domain containing 2C |
| chr10_+_38788992 | 2.08 |
ENSRNOT00000009727
|
Gdf9
|
growth differentiation factor 9 |
| chr4_-_180505916 | 2.01 |
ENSRNOT00000086465
|
AABR07062512.1
|
|
| chr1_+_172599610 | 2.00 |
ENSRNOT00000049903
|
Olr260
|
olfactory receptor 260 |
| chr19_+_24496569 | 1.97 |
ENSRNOT00000004982
|
Mgat4d
|
MGAT4 family, member D |
| chr10_+_61033113 | 1.97 |
ENSRNOT00000043016
|
Olr1513
|
olfactory receptor 1513 |
| chr1_-_173129038 | 1.90 |
ENSRNOT00000075693
|
LOC100912408
|
olfactory receptor 10A3-like |
| chr10_+_46783979 | 1.89 |
ENSRNOT00000005237
|
Gid4
|
GID complex subunit 4 |
| chr8_-_41445722 | 1.88 |
ENSRNOT00000011381
|
LOC171573
|
spleen protein 1 precursor |
| chr7_-_130120579 | 1.83 |
ENSRNOT00000044376
|
Mapk12
|
mitogen-activated protein kinase 12 |
| chr10_-_50574539 | 1.76 |
ENSRNOT00000034261
|
Cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| chr8_+_40410604 | 1.43 |
ENSRNOT00000049392
|
Olr1202
|
olfactory receptor 1202 |
| chr7_-_75569778 | 1.42 |
ENSRNOT00000040687
|
LOC362901
|
Ac1254 |
| chr10_-_41546346 | 1.36 |
ENSRNOT00000058627
|
LOC102552882
|
uncharacterized LOC102552882 |
| chr7_+_78649875 | 1.35 |
ENSRNOT00000006243
|
Dcstamp
|
dendrocyte expressed seven transmembrane protein |
| chr20_-_8574082 | 1.30 |
ENSRNOT00000048845
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr4_-_32087600 | 1.18 |
ENSRNOT00000013841
|
Sem1
|
SEM1, 26S proteasome complex subunit |
| chr7_-_142180997 | 1.06 |
ENSRNOT00000087632
|
Tfcp2
|
transcription factor CP2 |
| chr10_-_109966782 | 1.02 |
ENSRNOT00000078052
ENSRNOT00000075041 |
Rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_+_110749705 | 0.97 |
ENSRNOT00000084348
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr1_+_31436712 | 0.92 |
ENSRNOT00000038102
|
AC094217.1
|
|
| chr1_-_141533908 | 0.87 |
ENSRNOT00000020117
|
Mesp1
|
mesoderm posterior bHLH transcription factor 1 |
| chr8_+_41936959 | 0.77 |
ENSRNOT00000042601
|
Olr1222
|
olfactory receptor 1222 |
| chr1_+_172051215 | 0.71 |
ENSRNOT00000048370
|
Olr235
|
olfactory receptor 235 |
| chr6_-_11860001 | 0.67 |
ENSRNOT00000052280
|
H2afz
|
H2A histone family, member Z |
| chr10_+_11206226 | 0.54 |
ENSRNOT00000006979
|
Tfap4
|
transcription factor AP-4 |
| chr10_+_28449335 | 0.54 |
ENSRNOT00000029118
|
Atp10b
|
ATPase phospholipid transporting 10B (putative) |
| chr13_-_91737053 | 0.51 |
ENSRNOT00000083561
|
Fcer1a
|
Fc fragment of IgE receptor Ia |
| chr3_+_74411668 | 0.51 |
ENSRNOT00000045439
|
Olr526
|
olfactory receptor 526 |
| chr7_-_143453544 | 0.50 |
ENSRNOT00000034450
ENSRNOT00000083956 |
Krt1
Krt5
|
keratin 1 keratin 5 |
| chr11_-_55240241 | 0.35 |
ENSRNOT00000064614
|
Dppa4
|
developmental pluripotency associated 4 |
| chr1_-_226049929 | 0.33 |
ENSRNOT00000007320
|
Best1
|
bestrophin 1 |
| chr10_+_61160125 | 0.28 |
ENSRNOT00000074715
|
Olr1519
|
olfactory receptor 1519 |
| chr1_+_150225373 | 0.24 |
ENSRNOT00000051266
|
Olr30
|
olfactory receptor 30 |
| chr7_+_70697050 | 0.21 |
ENSRNOT00000045964
|
R3hdm2
|
R3H domain containing 2 |
| chr1_-_226353611 | 0.20 |
ENSRNOT00000037624
|
Dagla
|
diacylglycerol lipase, alpha |
| chr12_+_10255416 | 0.16 |
ENSRNOT00000092720
|
Gpr12
|
G protein-coupled receptor 12 |
| chr14_-_46153212 | 0.15 |
ENSRNOT00000079269
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr8_+_41987849 | 0.14 |
ENSRNOT00000073800
|
LOC100911999
|
olfactory receptor 143-like |
| chr6_+_129438158 | 0.03 |
ENSRNOT00000005953
|
Bdkrb1
|
bradykinin receptor B1 |
| chr9_-_81565416 | 0.01 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of T
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.9 | 47.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 11.1 | 88.9 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 9.7 | 48.4 | GO:0035106 | operant conditioning(GO:0035106) |
| 9.4 | 74.9 | GO:0042905 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 9.1 | 27.2 | GO:1904612 | response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) response to thyrotropin-releasing hormone(GO:1905225) |
| 8.4 | 25.2 | GO:0072237 | acetyl-CoA catabolic process(GO:0046356) ketone body catabolic process(GO:0046952) metanephric proximal tubule development(GO:0072237) |
| 7.5 | 22.5 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 6.3 | 25.3 | GO:0014028 | notochord formation(GO:0014028) |
| 6.0 | 47.8 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 5.9 | 35.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 5.3 | 15.9 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 4.0 | 12.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 3.8 | 11.4 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 3.7 | 14.8 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 2.9 | 2.9 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 2.8 | 11.3 | GO:0009597 | detection of virus(GO:0009597) |
| 2.8 | 27.8 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 2.6 | 15.4 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 2.2 | 13.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 2.1 | 14.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 2.1 | 10.4 | GO:0002018 | angiotensin maturation(GO:0002003) renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 2.0 | 26.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 1.8 | 20.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.7 | 8.6 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 1.6 | 16.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 1.6 | 19.5 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 1.6 | 8.0 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 1.6 | 18.8 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 1.6 | 42.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.5 | 14.7 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 1.5 | 42.1 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 1.4 | 4.2 | GO:1900222 | regulation of cell growth by extracellular stimulus(GO:0001560) positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 1.3 | 16.4 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 1.2 | 3.6 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 1.1 | 9.9 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 1.0 | 15.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 1.0 | 28.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 1.0 | 24.7 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 1.0 | 13.0 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.8 | 2.5 | GO:0015817 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.8 | 9.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.8 | 8.7 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.8 | 2.3 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.7 | 2.7 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.7 | 2.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.6 | 8.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.6 | 16.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.5 | 33.0 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.5 | 2.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.5 | 3.9 | GO:0061193 | taste bud development(GO:0061193) |
| 0.5 | 3.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.5 | 1.4 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.4 | 6.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.4 | 3.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.3 | 7.5 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.3 | 1.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.3 | 16.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.3 | 16.2 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.3 | 3.3 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.3 | 0.9 | GO:0060913 | mesodermal cell migration(GO:0008078) cardiac cell fate determination(GO:0060913) cardiac vascular smooth muscle cell differentiation(GO:0060947) cell migration involved in heart formation(GO:0060974) regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.2 | 6.2 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.2 | 5.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 3.3 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.2 | 3.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 3.8 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.2 | 0.5 | GO:0045399 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.2 | 3.7 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.5 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 8.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 3.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 1.8 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.2 | GO:0099542 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.1 | 5.2 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.1 | 22.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 3.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 2.6 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 1.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.4 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.8 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 3.3 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 23.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 2.8 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.7 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 4.4 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 18.8 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 2.0 | 16.3 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 1.4 | 4.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.3 | 37.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.8 | 7.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.7 | 2.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.7 | 6.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.6 | 26.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 8.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 3.4 | GO:0033290 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.3 | 16.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 3.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 33.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 4.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 20.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.3 | 3.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 3.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 73.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.2 | 5.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 3.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 2.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 36.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.2 | 24.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 4.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 14.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 2.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 8.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.7 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 6.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 15.2 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 5.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 7.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 15.9 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 25.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 56.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 8.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 2.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 7.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 13.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 1.8 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 142.9 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 5.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 43.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 3.8 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.4 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 14.8 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 88.7 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 41.1 | 123.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 11.1 | 88.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 8.4 | 25.2 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 5.6 | 27.8 | GO:0070404 | NADH binding(GO:0070404) |
| 5.3 | 15.9 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 4.9 | 14.8 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 3.1 | 15.4 | GO:0051185 | adenosine-diphosphatase activity(GO:0043262) coenzyme transporter activity(GO:0051185) |
| 2.9 | 35.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 2.8 | 47.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 2.2 | 8.7 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 1.7 | 8.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 1.7 | 5.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 1.4 | 13.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 1.4 | 4.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.4 | 42.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.3 | 6.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 1.3 | 16.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 1.2 | 57.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 1.0 | 39.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.9 | 26.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.8 | 2.5 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.8 | 16.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.8 | 8.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.7 | 12.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.6 | 25.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.6 | 7.5 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.6 | 2.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.6 | 3.9 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.5 | 9.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.5 | 22.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.5 | 3.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.4 | 24.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 10.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.3 | 8.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 2.7 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.3 | 29.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 5.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 1.8 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 3.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 3.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 27.2 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.2 | 8.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 7.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 3.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 4.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 14.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 3.7 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.2 | 36.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 11.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 14.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 6.5 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.1 | 18.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.5 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.1 | 5.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 3.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 3.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 7.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 5.9 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 12.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 12.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 4.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 14.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.0 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 2.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 4.0 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 47.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.9 | 35.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.8 | 25.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.8 | 38.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.6 | 4.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 24.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.4 | 9.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 69.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 4.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 6.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 4.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.8 | 102.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 2.7 | 27.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 2.4 | 47.8 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 1.4 | 26.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 1.4 | 25.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 1.1 | 13.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.0 | 11.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 1.0 | 16.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.8 | 22.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.6 | 4.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.5 | 7.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.5 | 14.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.4 | 4.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.3 | 3.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.2 | 16.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 8.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 3.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 14.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 2.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 1.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 8.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 5.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 9.6 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 8.0 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.1 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 3.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 3.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |