Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
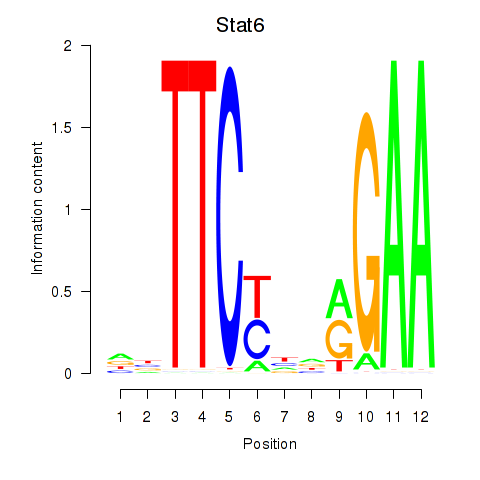
Results for Stat6
Z-value: 0.84
Transcription factors associated with Stat6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat6
|
ENSRNOG00000025023 | signal transducer and activator of transcription 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat6 | rn6_v1_chr7_+_70950231_70950231 | 0.14 | 1.4e-02 | Click! |
Activity profile of Stat6 motif
Sorted Z-values of Stat6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_63568464 | 30.76 |
ENSRNOT00000068494
|
AABR07052585.2
|
|
| chr16_-_18766174 | 27.40 |
ENSRNOT00000084813
|
Sftpd
|
surfactant protein D |
| chr18_-_61057365 | 22.89 |
ENSRNOT00000042352
|
Alpk2
|
alpha-kinase 2 |
| chr1_-_88066101 | 22.03 |
ENSRNOT00000079473
ENSRNOT00000027893 |
Ryr1
|
ryanodine receptor 1 |
| chr6_+_56625650 | 21.90 |
ENSRNOT00000008803
|
Meox2
|
mesenchyme homeobox 2 |
| chr1_-_198233588 | 18.22 |
ENSRNOT00000088473
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr1_-_198233215 | 17.81 |
ENSRNOT00000087928
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr4_-_148437961 | 17.52 |
ENSRNOT00000082907
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr1_-_143392532 | 16.50 |
ENSRNOT00000026089
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr2_+_199162745 | 16.40 |
ENSRNOT00000023488
|
Gja5
|
gap junction protein, alpha 5 |
| chr8_-_108777717 | 16.33 |
ENSRNOT00000034188
|
Il20rb
|
interleukin 20 receptor subunit beta |
| chr17_-_15467320 | 14.61 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chr7_-_60341264 | 14.09 |
ENSRNOT00000007747
|
Lyz2
|
lysozyme 2 |
| chr7_-_33793565 | 13.37 |
ENSRNOT00000065354
|
AABR07056633.1
|
|
| chr14_+_37113210 | 13.34 |
ENSRNOT00000089094
|
Sgcb
|
sarcoglycan, beta |
| chr15_+_83703791 | 12.78 |
ENSRNOT00000090637
|
Klf5
|
Kruppel-like factor 5 |
| chr13_-_70667658 | 12.58 |
ENSRNOT00000092521
|
Lamc1
|
laminin subunit gamma 1 |
| chrX_-_111887906 | 12.15 |
ENSRNOT00000085118
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr18_-_6782757 | 11.80 |
ENSRNOT00000068150
|
Aqp4
|
aquaporin 4 |
| chr8_-_114449956 | 10.68 |
ENSRNOT00000056414
|
Col6a6
|
collagen type VI alpha 6 chain |
| chr2_-_173836854 | 10.34 |
ENSRNOT00000074278
|
Wdr49
|
WD repeat domain 49 |
| chr9_+_66335492 | 10.33 |
ENSRNOT00000037555
|
RGD1562029
|
similar to KIAA2012 protein |
| chr15_+_41937880 | 9.75 |
ENSRNOT00000032514
|
Kcnrg
|
potassium channel regulator |
| chr16_+_29674793 | 9.61 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr7_+_59326518 | 9.55 |
ENSRNOT00000085231
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr4_+_78354335 | 9.52 |
ENSRNOT00000059167
|
Gimap7
|
GTPase, IMAP family member 7 |
| chr15_-_35417273 | 9.41 |
ENSRNOT00000083961
ENSRNOT00000041430 |
Gzmb
|
granzyme B |
| chr1_-_221281180 | 9.21 |
ENSRNOT00000028379
|
Cdc42ep2
|
CDC42 effector protein 2 |
| chr2_-_98610368 | 9.09 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr18_+_30840868 | 8.97 |
ENSRNOT00000027026
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr15_-_36114752 | 8.66 |
ENSRNOT00000045209
|
Gzmbl3
|
Granzyme B-like 3 |
| chr4_-_58881045 | 8.62 |
ENSRNOT00000087241
|
Podxl
|
podocalyxin-like |
| chr1_+_127802978 | 8.62 |
ENSRNOT00000055877
|
Adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr13_+_47454591 | 8.47 |
ENSRNOT00000005791
|
LOC498222
|
similar to specifically androgen-regulated protein |
| chr1_-_195096460 | 8.37 |
ENSRNOT00000077253
|
Snrpn
|
small nuclear ribonucleoprotein polypeptide N |
| chr15_-_36472327 | 8.23 |
ENSRNOT00000032601
ENSRNOT00000059660 |
LOC102553861
|
granzyme-like protein 1-like |
| chr4_-_133951264 | 7.98 |
ENSRNOT00000090506
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chrX_+_68891227 | 7.72 |
ENSRNOT00000009635
|
Efnb1
|
ephrin B1 |
| chr13_+_51022681 | 7.38 |
ENSRNOT00000078599
|
Chi3l1
|
chitinase 3 like 1 |
| chr1_-_217844957 | 7.20 |
ENSRNOT00000090653
|
Ano1
|
anoctamin 1 |
| chr9_+_111028824 | 6.98 |
ENSRNOT00000041418
ENSRNOT00000056457 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_+_171793782 | 6.86 |
ENSRNOT00000081351
|
Olfml1
|
olfactomedin-like 1 |
| chr8_-_114940163 | 6.71 |
ENSRNOT00000083303
ENSRNOT00000086232 |
Alas1
|
5'-aminolevulinate synthase 1 |
| chr5_-_163231494 | 6.70 |
ENSRNOT00000085929
ENSRNOT00000022968 |
Tnfrsf8
|
TNF receptor superfamily member 8 |
| chr7_-_70467915 | 6.65 |
ENSRNOT00000088995
|
Slc26a10
|
solute carrier family 26, member 10 |
| chr7_+_97559841 | 6.38 |
ENSRNOT00000007326
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr19_-_22281778 | 6.37 |
ENSRNOT00000049624
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr3_+_139894331 | 6.24 |
ENSRNOT00000064695
|
Rin2
|
Ras and Rab interactor 2 |
| chr5_+_139597731 | 5.85 |
ENSRNOT00000072427
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr18_+_30913842 | 5.84 |
ENSRNOT00000026947
|
Pcdhgc3
|
protocadherin gamma subfamily C, 3 |
| chr15_+_4209703 | 5.80 |
ENSRNOT00000082236
|
Ppp3cb
|
protein phosphatase 3 catalytic subunit beta |
| chr5_-_166116516 | 5.56 |
ENSRNOT00000079919
ENSRNOT00000080888 |
Kif1b
|
kinesin family member 1B |
| chr1_+_282715344 | 5.55 |
ENSRNOT00000074399
|
LOC100910259
|
liver carboxylesterase-like |
| chr12_+_40553741 | 5.50 |
ENSRNOT00000057396
|
LOC100360238
|
rCG21419-like |
| chr1_+_198199622 | 5.46 |
ENSRNOT00000026688
|
Gdpd3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr20_+_5057701 | 5.40 |
ENSRNOT00000001119
|
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C |
| chr4_-_77706994 | 5.30 |
ENSRNOT00000038517
|
Zfp777
|
zinc finger protein 777 |
| chr18_+_29506128 | 5.17 |
ENSRNOT00000003878
|
Slc35a4
|
solute carrier family 35, member A4 |
| chr8_-_53816447 | 5.09 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr13_+_88265331 | 5.05 |
ENSRNOT00000031190
|
Ccdc190
|
coiled-coil domain containing 190 |
| chrX_+_20351486 | 4.76 |
ENSRNOT00000093675
ENSRNOT00000047444 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr8_-_129919120 | 4.66 |
ENSRNOT00000074423
|
Ulk4
|
unc-51 like kinase 4 |
| chr4_-_10329241 | 4.53 |
ENSRNOT00000017232
|
Fgl2
|
fibrinogen-like 2 |
| chr15_+_3938075 | 4.16 |
ENSRNOT00000065644
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr1_+_128606770 | 4.15 |
ENSRNOT00000073355
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chrX_+_20423401 | 4.08 |
ENSRNOT00000093162
|
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr1_+_64114721 | 4.08 |
ENSRNOT00000080466
|
Tmc4
|
transmembrane channel-like 4 |
| chr13_-_109580271 | 4.02 |
ENSRNOT00000088326
|
Flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr19_+_43163129 | 4.01 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr10_-_89130339 | 3.87 |
ENSRNOT00000027640
|
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr2_+_211337271 | 3.60 |
ENSRNOT00000045155
|
Cox6b1
|
cytochrome c oxidase subunit 6B1 |
| chr18_+_30820321 | 3.52 |
ENSRNOT00000060472
|
Pcdhga3
|
protocadherin gamma subfamily A, 3 |
| chr1_+_221710670 | 3.48 |
ENSRNOT00000064798
|
Map4k2
|
mitogen activated protein kinase kinase kinase kinase 2 |
| chr3_-_91195981 | 3.24 |
ENSRNOT00000056935
|
RGD1309730
|
similar to RIKEN cDNA B230118H07 |
| chr3_+_7109920 | 3.18 |
ENSRNOT00000084092
ENSRNOT00000013809 |
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr15_-_12513931 | 3.07 |
ENSRNOT00000010103
|
Atxn7
|
ataxin 7 |
| chr4_-_157266018 | 3.05 |
ENSRNOT00000019570
|
LOC100911713
|
protein C10-like |
| chr14_+_7618022 | 2.99 |
ENSRNOT00000088508
ENSRNOT00000002819 |
Slc10a6
|
solute carrier family 10 member 6 |
| chr5_-_135561914 | 2.93 |
ENSRNOT00000023178
|
Mmachc
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr9_-_50762082 | 2.74 |
ENSRNOT00000015492
|
Mettl21c
|
methyltransferase like 21C |
| chr1_-_6970040 | 2.62 |
ENSRNOT00000016273
|
Utrn
|
utrophin |
| chr10_-_6870011 | 2.58 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
| chr15_-_35166211 | 2.57 |
ENSRNOT00000059662
|
LOC691670
|
similar to natural killer cell protease 7 |
| chr1_-_103323476 | 2.53 |
ENSRNOT00000019051
|
Mrgprx3
|
MAS related GPR family member X3 |
| chr13_+_48790951 | 2.36 |
ENSRNOT00000090233
|
Elk4
|
ELK4, ETS transcription factor |
| chr13_+_27032048 | 2.23 |
ENSRNOT00000031789
|
Serpinb13
|
serpin family B member 13 |
| chr17_+_47241017 | 2.21 |
ENSRNOT00000092193
|
Gpr141
|
G protein-coupled receptor 141 |
| chr17_+_23116661 | 2.20 |
ENSRNOT00000067374
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr2_+_189629297 | 2.18 |
ENSRNOT00000049810
|
Dennd4b
|
DENN domain containing 4B |
| chr4_-_23119005 | 2.12 |
ENSRNOT00000048061
|
Steap4
|
STEAP4 metalloreductase |
| chr13_+_48790767 | 2.02 |
ENSRNOT00000087504
|
Elk4
|
ELK4, ETS transcription factor |
| chr19_-_38796 | 2.01 |
ENSRNOT00000077592
|
AABR07042565.1
|
|
| chr1_-_172030164 | 1.96 |
ENSRNOT00000026807
|
Ovch2
|
ovochymase 2 |
| chr7_+_143897014 | 1.94 |
ENSRNOT00000017314
|
Espl1
|
extra spindle pole bodies like 1, separase |
| chr14_+_107750162 | 1.87 |
ENSRNOT00000012767
|
Zrsr1
|
zinc finger (CCCH type), RNA binding motif and serine/arginine rich 1 |
| chr7_-_143473697 | 1.82 |
ENSRNOT00000055336
ENSRNOT00000083027 |
Krt77
|
keratin 77 |
| chr20_+_48881194 | 1.80 |
ENSRNOT00000000304
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr4_+_183896303 | 1.77 |
ENSRNOT00000055437
|
RGD1309621
|
similar to hypothetical protein FLJ10652 |
| chr19_-_37725623 | 1.72 |
ENSRNOT00000024197
|
Gfod2
|
glucose-fructose oxidoreductase domain containing 2 |
| chr2_+_149899836 | 1.71 |
ENSRNOT00000086481
|
RGD1560324
|
similar to hypothetical protein C130079G13 |
| chr13_+_99387950 | 1.70 |
ENSRNOT00000004815
|
RGD1564463
|
similar to Mdes protein |
| chr14_-_28967980 | 1.68 |
ENSRNOT00000048175
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr11_+_61970976 | 1.67 |
ENSRNOT00000078921
|
Tigit
|
T cell immunoreceptor with Ig and ITIM domains |
| chr15_-_35195725 | 1.49 |
ENSRNOT00000044840
|
Gzmf
|
granzyme F |
| chr3_-_74906989 | 1.49 |
ENSRNOT00000071001
|
Olr545
|
olfactory receptor 545 |
| chr10_+_106712127 | 1.48 |
ENSRNOT00000040629
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr11_-_54344615 | 1.47 |
ENSRNOT00000090307
|
Myh15
|
myosin, heavy chain 15 |
| chr8_-_49158971 | 1.44 |
ENSRNOT00000020573
|
Kmt2a
|
lysine methyltransferase 2A |
| chr1_-_193700673 | 1.42 |
ENSRNOT00000071314
|
Paip2l1
|
polyadenylate-binding protein-interacting protein 2-like 1 |
| chr11_-_78341349 | 1.42 |
ENSRNOT00000068116
|
Tp63
|
tumor protein p63 |
| chr20_+_7818289 | 1.39 |
ENSRNOT00000042539
|
Ppard
|
peroxisome proliferator-activated receptor delta |
| chr20_+_2057878 | 1.38 |
ENSRNOT00000051480
|
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr2_-_90568486 | 1.38 |
ENSRNOT00000059380
|
LOC100910852
|
uncharacterized LOC100910852 |
| chr20_+_1764794 | 1.34 |
ENSRNOT00000075084
|
Olr1736
|
olfactory receptor 1736 |
| chr13_+_90184569 | 1.33 |
ENSRNOT00000082393
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr19_-_10581842 | 1.31 |
ENSRNOT00000021639
|
Polr2c
|
RNA polymerase II subunit C |
| chr3_-_118427851 | 1.29 |
ENSRNOT00000055995
|
Fam227b
|
family with sequence similarity 227, member B |
| chr10_-_56368360 | 1.29 |
ENSRNOT00000019674
|
Slc35g3
|
solute carrier family 35, member G3 |
| chr13_-_91776397 | 1.26 |
ENSRNOT00000073147
|
Mptx1
|
mucosal pentraxin 1 |
| chr20_+_2057689 | 1.22 |
ENSRNOT00000086007
|
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr9_+_20279938 | 1.20 |
ENSRNOT00000075612
|
Grcc10
|
gene rich cluster, C10 gene |
| chr8_-_55696601 | 1.18 |
ENSRNOT00000016127
|
RGD1562914
|
RGD1562914 |
| chr11_-_78327282 | 1.18 |
ENSRNOT00000002636
|
Tp63
|
tumor protein p63 |
| chr8_+_113939886 | 1.17 |
ENSRNOT00000079562
|
Aste1
|
asteroid homolog 1 (Drosophila) |
| chr1_+_260153645 | 1.11 |
ENSRNOT00000054717
|
Zfp518a
|
zinc finger protein 518A |
| chr1_-_124803363 | 1.06 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr8_-_36374673 | 1.04 |
ENSRNOT00000013763
|
Dcps
|
decapping enzyme, scavenger |
| chr4_+_1841955 | 1.02 |
ENSRNOT00000019271
|
Olr1096
|
olfactory receptor 1096 |
| chr7_+_9262982 | 0.99 |
ENSRNOT00000047647
|
Olr1063
|
olfactory receptor 1063 |
| chr1_-_149683357 | 0.96 |
ENSRNOT00000077975
|
Olr17
|
olfactory receptor 17 |
| chr3_+_21698032 | 0.94 |
ENSRNOT00000013378
|
Rabgap1
|
RAB GTPase activating protein 1 |
| chr8_+_43606676 | 0.90 |
ENSRNOT00000083262
|
Olr1319
|
olfactory receptor 1319 |
| chr20_+_1736377 | 0.89 |
ENSRNOT00000047035
|
Olr1734
|
olfactory receptor 1734 |
| chr11_+_60054408 | 0.88 |
ENSRNOT00000091756
|
Abhd10
|
abhydrolase domain containing 10 |
| chr15_+_61826937 | 0.79 |
ENSRNOT00000084005
ENSRNOT00000079417 |
Elf1
|
E74-like factor 1 |
| chr9_-_63291350 | 0.72 |
ENSRNOT00000058831
|
Hsfy2
|
heat shock transcription factor, Y linked 2 |
| chr1_+_63184734 | 0.71 |
ENSRNOT00000078889
|
Vom1r110
|
vomeronasal 1 receptor 110 |
| chr8_-_47094352 | 0.69 |
ENSRNOT00000048347
|
Grik4
|
glutamate ionotropic receptor kainate type subunit 4 |
| chr13_-_47438360 | 0.69 |
ENSRNOT00000067445
|
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr16_-_7412150 | 0.67 |
ENSRNOT00000047252
|
RGD1564325
|
similar to ribosomal protein S24 |
| chr8_+_41235156 | 0.66 |
ENSRNOT00000071644
|
LOC100911958
|
olfactory receptor 147-like |
| chr15_-_36158195 | 0.64 |
ENSRNOT00000091636
|
Olr1288
|
olfactory receptor 1288 |
| chr3_-_76718684 | 0.61 |
ENSRNOT00000050173
|
Olr631
|
olfactory receptor 631 |
| chr5_+_135562034 | 0.61 |
ENSRNOT00000056967
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr8_+_45797315 | 0.58 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chr16_+_2634603 | 0.57 |
ENSRNOT00000019113
|
Hesx1
|
HESX homeobox 1 |
| chr15_+_35685009 | 0.44 |
ENSRNOT00000085764
|
LOC100911127
|
olfactory receptor 144-like |
| chr11_+_80319177 | 0.43 |
ENSRNOT00000036777
|
Rtp2
|
receptor (chemosensory) transporter protein 2 |
| chr15_+_34282936 | 0.42 |
ENSRNOT00000026364
|
Irf9
|
interferon regulatory factor 9 |
| chr15_-_34392066 | 0.37 |
ENSRNOT00000027315
|
Tgm1
|
transglutaminase 1 |
| chr1_+_218076116 | 0.35 |
ENSRNOT00000028374
|
Oraov1
|
oral cancer overexpressed 1 |
| chr1_-_168710460 | 0.33 |
ENSRNOT00000047399
|
Olr114
|
olfactory receptor 114 |
| chr1_+_88582641 | 0.33 |
ENSRNOT00000028187
|
Zfp382
|
zinc finger protein 382 |
| chr20_+_1749716 | 0.32 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr8_+_62317265 | 0.30 |
ENSRNOT00000083418
ENSRNOT00000091204 |
Fam219b
|
family with sequence similarity 219, member B |
| chr10_-_78993045 | 0.29 |
ENSRNOT00000043005
|
LOC100363469
|
ribosomal protein S24-like |
| chr14_+_76657311 | 0.26 |
ENSRNOT00000076730
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr19_-_10620671 | 0.23 |
ENSRNOT00000021842
|
Ccl17
|
C-C motif chemokine ligand 17 |
| chr8_-_43235208 | 0.18 |
ENSRNOT00000060095
|
Olr1304
|
olfactory receptor 1304 |
| chr7_-_5106708 | 0.16 |
ENSRNOT00000046001
|
Olr892
|
olfactory receptor 892 |
| chr3_-_77893647 | 0.11 |
ENSRNOT00000080803
|
Olr678
|
olfactory receptor 678 |
| chr1_-_170841363 | 0.10 |
ENSRNOT00000046171
|
Olr213
|
olfactory receptor 213 |
| chr1_-_226606344 | 0.05 |
ENSRNOT00000028079
|
Tmem216
|
transmembrane protein 216 |
| chrX_-_135419704 | 0.03 |
ENSRNOT00000044435
|
Zfp280c
|
zinc finger protein 280C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 27.4 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 5.5 | 16.4 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 5.4 | 16.3 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 4.4 | 17.5 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 4.4 | 21.9 | GO:0001757 | somite specification(GO:0001757) |
| 3.1 | 22.0 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 3.1 | 9.4 | GO:0060545 | positive regulation of necroptotic process(GO:0060545) |
| 2.9 | 11.8 | GO:0060354 | negative regulation of cell adhesion molecule production(GO:0060354) |
| 2.9 | 8.8 | GO:2000682 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 2.6 | 36.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 2.2 | 6.7 | GO:0032759 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 1.9 | 5.8 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 1.8 | 21.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 1.7 | 7.0 | GO:0031179 | peptide modification(GO:0031179) |
| 1.3 | 4.0 | GO:0097037 | heme export(GO:0097037) |
| 1.3 | 13.3 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 1.2 | 12.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 1.2 | 3.5 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 1.1 | 6.7 | GO:0010045 | response to nickel cation(GO:0010045) response to platinum ion(GO:0070541) |
| 1.1 | 5.6 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.8 | 7.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.8 | 9.8 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.7 | 5.2 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.7 | 9.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.7 | 12.6 | GO:0070831 | hemidesmosome assembly(GO:0031581) basement membrane assembly(GO:0070831) |
| 0.6 | 2.6 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.5 | 1.4 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.5 | 14.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.4 | 1.3 | GO:0002277 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.4 | 7.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.4 | 2.9 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.4 | 6.7 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.4 | 3.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.3 | 4.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.3 | 2.2 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.3 | 2.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 7.7 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.3 | 0.9 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.3 | 3.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 | 2.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 4.7 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.2 | 16.0 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.2 | 1.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 3.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 10.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.2 | 4.4 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.2 | 4.5 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 0.6 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 1.9 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 1.5 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 6.4 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 1.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 1.0 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.7 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.3 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.1 | 1.7 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.1 | 3.6 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 5.4 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 10.2 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 1.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.4 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.0 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 1.7 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.0 | 0.4 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 1.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.6 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 2.4 | 22.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 2.3 | 14.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 2.2 | 13.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 2.1 | 36.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 1.8 | 17.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 1.2 | 9.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 1.0 | 5.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.9 | 8.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.8 | 6.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.8 | 16.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.7 | 27.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.7 | 8.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.6 | 5.6 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.3 | 11.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.3 | 1.9 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 2.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 3.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 4.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 8.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 35.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 7.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 22.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 1.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 2.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.5 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 5.5 | 22.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 5.4 | 16.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 5.1 | 36.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 4.1 | 16.4 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 2.2 | 6.7 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 1.8 | 7.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 1.7 | 7.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.7 | 12.1 | GO:0043426 | MRF binding(GO:0043426) |
| 1.5 | 9.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 1.5 | 5.8 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.4 | 12.6 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 1.2 | 7.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 1.1 | 27.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 1.1 | 14.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.9 | 5.6 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.8 | 11.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.8 | 6.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.7 | 8.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.6 | 3.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.6 | 3.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.6 | 1.7 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.6 | 3.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.5 | 6.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.5 | 21.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.5 | 6.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.4 | 4.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.4 | 5.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.3 | 0.9 | GO:0102390 | mycophenolic acid acyl-glucuronide esterase activity(GO:0102390) |
| 0.3 | 1.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.3 | 5.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 7.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 1.9 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 4.0 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 1.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 4.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 2.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 9.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 30.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 2.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 3.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 5.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 1.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.7 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 1.4 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.1 | 2.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 2.7 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 2.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 22.3 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 16.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 2.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 8.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 4.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 6.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.9 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.7 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.6 | 22.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.5 | 36.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.4 | 19.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.4 | 6.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 14.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 7.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.3 | 10.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 4.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 7.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 5.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 6.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 11.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 2.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 10.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 3.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 4.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 14.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 1.0 | 11.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.9 | 36.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.9 | 16.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.4 | 6.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 6.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 5.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 9.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 3.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 3.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.8 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 10.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 12.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 4.0 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 3.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |