Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
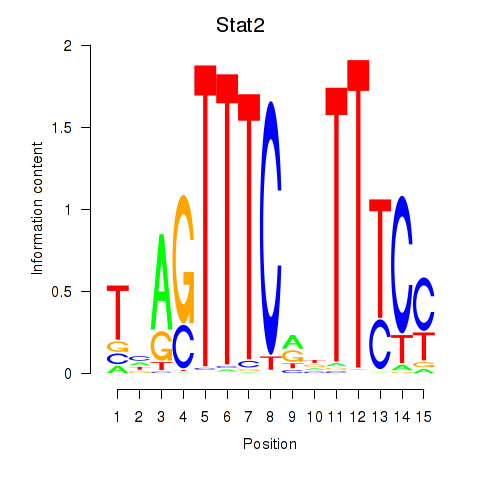
Results for Stat2
Z-value: 2.04
Transcription factors associated with Stat2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat2
|
ENSRNOG00000031081 | signal transducer and activator of transcription 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat2 | rn6_v1_chr7_+_2691369_2691369 | 0.74 | 2.0e-56 | Click! |
Activity profile of Stat2 motif
Sorted Z-values of Stat2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_58198782 | 110.28 |
ENSRNOT00000023951
|
Ccl21
|
C-C motif chemokine ligand 21 |
| chr15_+_18710492 | 101.52 |
ENSRNOT00000012532
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr20_+_3990820 | 86.58 |
ENSRNOT00000000528
|
Psmb8
|
proteasome subunit beta 8 |
| chr10_-_70744315 | 75.67 |
ENSRNOT00000014865
|
Ccl5
|
C-C motif chemokine ligand 5 |
| chr4_+_153805993 | 72.28 |
ENSRNOT00000056174
|
Usp18
|
ubiquitin specific peptidase 18 |
| chr3_-_171286413 | 71.44 |
ENSRNOT00000008365
ENSRNOT00000081036 |
Zbp1
|
Z-DNA binding protein 1 |
| chr10_-_70342411 | 70.88 |
ENSRNOT00000076269
ENSRNOT00000076477 |
Slfn13
|
schlafen family member 13 |
| chr13_+_89385859 | 69.12 |
ENSRNOT00000047434
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr5_-_58183017 | 68.40 |
ENSRNOT00000020982
|
Ccl19
|
C-C motif chemokine ligand 19 |
| chr20_-_27682861 | 66.48 |
ENSRNOT00000057317
|
Fam26f
|
family with sequence similarity 26, member F |
| chr3_-_176744377 | 65.63 |
ENSRNOT00000017787
|
Helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr4_-_163403653 | 64.73 |
ENSRNOT00000088151
|
Klrk1
|
killer cell lectin like receptor K1 |
| chrX_-_105390580 | 64.45 |
ENSRNOT00000077547
|
Btk
|
Bruton tyrosine kinase |
| chr13_+_89386023 | 63.05 |
ENSRNOT00000086223
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr1_-_214252456 | 62.26 |
ENSRNOT00000023504
|
Irf7
|
interferon regulatory factor 7 |
| chr20_+_3979035 | 58.48 |
ENSRNOT00000000529
|
Tap1
|
transporter 1, ATP binding cassette subfamily B member |
| chr2_+_209097927 | 55.74 |
ENSRNOT00000023807
|
Dennd2d
|
DENN domain containing 2D |
| chr1_+_215628785 | 55.15 |
ENSRNOT00000054864
|
Lsp1
|
lymphocyte-specific protein 1 |
| chr4_+_162493908 | 55.13 |
ENSRNOT00000072064
|
Clec2d
|
C-type lectin domain family 2, member D |
| chr12_+_41200718 | 54.63 |
ENSRNOT00000038426
ENSRNOT00000048450 ENSRNOT00000067176 |
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr18_+_55576239 | 52.95 |
ENSRNOT00000050063
|
LOC100910979
|
interferon-inducible GTPase 1-like |
| chr9_-_92616165 | 52.38 |
ENSRNOT00000056995
|
Sp110
|
SP110 nuclear body protein |
| chr1_+_279867034 | 50.85 |
ENSRNOT00000024164
|
Pnliprp1
|
pancreatic lipase-related protein 1 |
| chr2_+_248178389 | 50.45 |
ENSRNOT00000037339
|
Gbp5
|
guanylate binding protein 5 |
| chr20_-_3793985 | 49.17 |
ENSRNOT00000049540
ENSRNOT00000086293 |
RT1-CE16
|
RT1 class I, locus CE16 |
| chr2_-_190100276 | 48.32 |
ENSRNOT00000015351
|
S100a9
|
S100 calcium binding protein A9 |
| chr11_-_37914983 | 47.63 |
ENSRNOT00000039876
|
Mx1
|
myxovirus (influenza virus) resistance 1 |
| chr16_-_19942343 | 45.78 |
ENSRNOT00000087162
ENSRNOT00000091906 |
Bst2
|
bone marrow stromal cell antigen 2 |
| chr20_+_4855829 | 45.63 |
ENSRNOT00000001110
|
LOC103694380
|
tumor necrosis factor-like |
| chr12_+_41341417 | 45.54 |
ENSRNOT00000072024
ENSRNOT00000091460 |
Oas2
|
2'-5' oligoadenylate synthetase 2 |
| chr13_+_93684437 | 44.81 |
ENSRNOT00000005005
|
Kmo
|
kynurenine 3-monooxygenase |
| chr4_+_102351036 | 44.40 |
ENSRNOT00000079277
|
AABR07060994.1
|
|
| chrX_+_54390733 | 43.42 |
ENSRNOT00000004977
|
RGD1565785
|
similar to chromosome X open reading frame 21 |
| chr20_-_3978845 | 43.09 |
ENSRNOT00000000532
|
Psmb9
|
proteasome subunit beta 9 |
| chr1_+_169433539 | 42.69 |
ENSRNOT00000055210
|
Trim34
|
tripartite motif-containing 34 |
| chr10_+_76343847 | 42.48 |
ENSRNOT00000055674
|
Trim25
|
tripartite motif-containing 25 |
| chr6_+_127327959 | 41.98 |
ENSRNOT00000012296
|
Ifi27
|
interferon, alpha-inducible protein 27 |
| chr10_-_66229311 | 41.87 |
ENSRNOT00000016897
|
Lgals5
|
lectin, galactose binding, soluble 5 |
| chr10_+_43613931 | 41.66 |
ENSRNOT00000039335
|
Igtp
|
interferon gamma induced GTPase |
| chr11_+_38035611 | 41.37 |
ENSRNOT00000087603
ENSRNOT00000002695 |
Mx2
|
MX dynamin like GTPase 2 |
| chr10_-_5253336 | 41.31 |
ENSRNOT00000085310
|
Ciita
|
class II, major histocompatibility complex, transactivator |
| chr10_+_94944436 | 40.94 |
ENSRNOT00000078968
|
Milr1
|
mast cell immunoglobulin-like receptor 1 |
| chr4_+_67165883 | 40.71 |
ENSRNOT00000012280
|
Rab19
|
RAB19, member RAS oncogene family |
| chr11_+_67757928 | 40.39 |
ENSRNOT00000039215
|
Dtx3l
|
deltex E3 ubiquitin ligase 3L |
| chr16_-_85306366 | 40.23 |
ENSRNOT00000089650
|
Tnfsf13b
|
tumor necrosis factor superfamily member 13b |
| chr11_+_68105369 | 40.11 |
ENSRNOT00000046888
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr10_+_31880918 | 39.66 |
ENSRNOT00000059448
|
Timd4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr7_-_117300662 | 39.36 |
ENSRNOT00000006376
|
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr1_+_252906234 | 39.14 |
ENSRNOT00000031025
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr17_+_9236249 | 38.98 |
ENSRNOT00000015878
|
Tifab
|
TIFA inhibitor |
| chr15_+_34282936 | 38.83 |
ENSRNOT00000026364
|
Irf9
|
interferon regulatory factor 9 |
| chr10_-_107424710 | 37.88 |
ENSRNOT00000004320
|
Lgals3bp
|
galectin 3 binding protein |
| chr8_+_116754178 | 37.61 |
ENSRNOT00000068295
ENSRNOT00000084429 |
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr10_-_88163712 | 36.95 |
ENSRNOT00000005382
ENSRNOT00000084493 |
Krt17
|
keratin 17 |
| chr2_-_243224883 | 36.16 |
ENSRNOT00000014139
|
Dapp1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides 1 |
| chr15_+_60084918 | 36.10 |
ENSRNOT00000012632
|
Epsti1
|
epithelial stromal interaction 1 |
| chr20_+_3146856 | 35.89 |
ENSRNOT00000050159
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr11_+_67082193 | 35.61 |
ENSRNOT00000003129
|
Cd86
|
CD86 molecule |
| chr1_+_221538104 | 35.40 |
ENSRNOT00000028527
|
Batf2
|
basic leucine zipper ATF-like transcription factor 2 |
| chr8_-_63034226 | 35.09 |
ENSRNOT00000043434
|
Pml
|
promyelocytic leukemia |
| chr5_-_173626248 | 34.80 |
ENSRNOT00000039263
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr13_-_83917001 | 34.41 |
ENSRNOT00000004387
ENSRNOT00000082422 |
Rcsd1
|
RCSD domain containing 1 |
| chr3_-_37480984 | 34.09 |
ENSRNOT00000030373
|
Nmi
|
N-myc (and STAT) interactor |
| chr12_+_41155497 | 33.17 |
ENSRNOT00000041741
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr16_-_85305782 | 32.81 |
ENSRNOT00000067511
ENSRNOT00000076737 |
Tnfsf13b
|
tumor necrosis factor superfamily member 13b |
| chr1_-_252922486 | 32.58 |
ENSRNOT00000076802
|
Ifit1bl
|
interferon-induced protein with tetratricopeptide repeats 1B-like |
| chr2_-_256915563 | 32.28 |
ENSRNOT00000084873
ENSRNOT00000029990 |
Ifi44
|
interferon-induced protein 44 |
| chr17_-_32661865 | 31.99 |
ENSRNOT00000022194
|
Serpinb9
|
serpin family B member 9 |
| chr10_-_4257868 | 31.59 |
ENSRNOT00000035886
|
Tnfrsf17
|
TNF receptor superfamily member 17 |
| chr1_-_167708685 | 31.20 |
ENSRNOT00000092857
ENSRNOT00000024992 |
Trim21
|
tripartite motif-containing 21 |
| chr12_+_42343123 | 30.66 |
ENSRNOT00000043279
|
Oas1i
|
2 ' -5 ' oligoadenylate synthetase 1I |
| chr20_-_4542073 | 30.50 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr3_-_72171078 | 30.05 |
ENSRNOT00000009817
|
Serping1
|
serpin family G member 1 |
| chr15_+_34256071 | 30.02 |
ENSRNOT00000025887
|
Psme1
|
proteasome activator subunit 1 |
| chr10_+_89358376 | 29.15 |
ENSRNOT00000028067
|
Ifi35
|
interferon-induced protein 35 |
| chr3_+_161519743 | 28.31 |
ENSRNOT00000055148
|
Cd40
|
CD40 molecule |
| chr20_-_3166569 | 28.18 |
ENSRNOT00000091390
|
AABR07044362.1
|
|
| chr11_+_38035450 | 28.07 |
ENSRNOT00000083067
|
Mx2
|
MX dynamin like GTPase 2 |
| chr9_-_26932201 | 27.94 |
ENSRNOT00000017081
|
Mcm3
|
minichromosome maintenance complex component 3 |
| chr10_+_104377748 | 27.83 |
ENSRNOT00000080714
|
Llgl2
|
LLGL2, scribble cell polarity complex component |
| chr10_+_58860940 | 27.51 |
ENSRNOT00000056551
ENSRNOT00000074523 |
XAF1
|
XIAP associated factor-1 |
| chr4_+_179398621 | 26.69 |
ENSRNOT00000049474
ENSRNOT00000067506 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr10_+_108527740 | 26.58 |
ENSRNOT00000044983
|
Rnf213
|
ring finger protein 213 |
| chr15_+_36809361 | 26.37 |
ENSRNOT00000076667
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr1_-_224974203 | 25.85 |
ENSRNOT00000065694
|
Tmem179b
|
transmembrane protein 179B |
| chr4_-_66062089 | 25.81 |
ENSRNOT00000018782
|
Zc3hav1
|
zinc finger CCCH-type containing, antiviral 1 |
| chr10_+_64737022 | 25.68 |
ENSRNOT00000017071
ENSRNOT00000093232 ENSRNOT00000017042 ENSRNOT00000093244 |
Lgals9
|
galectin 9 |
| chr11_-_14304603 | 25.66 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr20_+_3167079 | 25.38 |
ENSRNOT00000001035
|
RT1-N3
|
RT1 class Ib, locus N3 |
| chr14_+_17210733 | 25.35 |
ENSRNOT00000003075
|
Cxcl10
|
C-X-C motif chemokine ligand 10 |
| chr9_-_54351339 | 25.30 |
ENSRNOT00000080522
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr10_-_57064600 | 25.00 |
ENSRNOT00000032926
|
Cxcl16
|
C-X-C motif chemokine ligand 16 |
| chr4_-_21920651 | 24.71 |
ENSRNOT00000066211
|
Tmem243
|
transmembrane protein 243 |
| chr7_+_11878943 | 24.37 |
ENSRNOT00000025634
|
Mob3a
|
MOB kinase activator 3A |
| chr10_+_90376933 | 24.29 |
ENSRNOT00000028557
|
Grn
|
granulin precursor |
| chr8_-_85803433 | 24.13 |
ENSRNOT00000081544
ENSRNOT00000073286 |
Mb21d1
|
Mab-21 domain containing 1 |
| chr20_-_5476193 | 23.66 |
ENSRNOT00000044975
|
Tapbp
|
TAP binding protein |
| chr2_+_248276709 | 23.21 |
ENSRNOT00000068683
|
Gbp2
|
guanylate binding protein 2 |
| chr5_+_113725717 | 23.02 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr17_-_43640387 | 22.32 |
ENSRNOT00000087731
|
Hist1h1c
|
histone cluster 1 H1 family member c |
| chr10_-_34221928 | 22.24 |
ENSRNOT00000045545
|
Irgm
|
immunity-related GTPase M |
| chr10_+_90377103 | 22.03 |
ENSRNOT00000040472
|
Grn
|
granulin precursor |
| chr3_-_48604097 | 21.08 |
ENSRNOT00000009620
|
Ifih1
|
interferon induced with helicase C domain 1 |
| chr10_-_110431792 | 20.11 |
ENSRNOT00000054922
|
LOC619574
|
hypothetical protein LOC619574 |
| chr20_+_3269284 | 19.92 |
ENSRNOT00000089044
|
RT1-T24-1
|
RT1 class I, locus T24, gene 1 |
| chr5_-_62153762 | 19.40 |
ENSRNOT00000066001
ENSRNOT00000086962 |
Trim14
|
tripartite motif-containing 14 |
| chr20_+_3242470 | 19.26 |
ENSRNOT00000078110
ENSRNOT00000041502 |
RT1-T24-2
RT1-T24-1
|
RT1 class I, locus T24, gene 2 RT1 class I, locus T24, gene 1 |
| chr19_-_55367353 | 19.25 |
ENSRNOT00000091139
|
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr17_+_53956203 | 19.21 |
ENSRNOT00000022483
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr13_+_99220510 | 19.17 |
ENSRNOT00000004519
|
Tmem63a
|
transmembrane protein 63a |
| chr7_-_140600818 | 18.30 |
ENSRNOT00000082375
|
Lmbr1l
|
limb development membrane protein 1-like |
| chr10_-_88611105 | 18.26 |
ENSRNOT00000024718
|
Dhx58
|
DEXH-box helicase 58 |
| chr1_+_100832324 | 18.08 |
ENSRNOT00000056364
|
Il4i1
|
interleukin 4 induced 1 |
| chr15_+_49010492 | 17.92 |
ENSRNOT00000024527
|
Nuggc
|
nuclear GTPase, germinal center associated |
| chr1_+_247562202 | 17.54 |
ENSRNOT00000021614
|
Pdcd1lg2
|
programmed cell death 1 ligand 2 |
| chr10_-_16046033 | 17.14 |
ENSRNOT00000089460
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr5_+_117586103 | 16.89 |
ENSRNOT00000084640
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr12_-_47456894 | 16.81 |
ENSRNOT00000001570
|
Oasl
|
2'-5'-oligoadenylate synthetase-like |
| chr4_-_66899914 | 16.74 |
ENSRNOT00000011481
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr4_-_157743199 | 16.58 |
ENSRNOT00000038178
|
Tapbpl
|
TAP binding protein-like |
| chr18_+_55685613 | 16.41 |
ENSRNOT00000040116
|
MGC105567
|
similar to cDNA sequence BC023105 |
| chr1_+_166125474 | 16.23 |
ENSRNOT00000091822
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr4_-_81241152 | 16.08 |
ENSRNOT00000015325
ENSRNOT00000015152 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr2_-_210550490 | 16.01 |
ENSRNOT00000081835
ENSRNOT00000025222 ENSRNOT00000086403 |
Csf1
|
colony stimulating factor 1 |
| chr3_+_114087287 | 15.89 |
ENSRNOT00000023017
|
B2m
|
beta-2 microglobulin |
| chr6_+_135866739 | 15.88 |
ENSRNOT00000013460
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr1_-_53520788 | 15.24 |
ENSRNOT00000060121
|
Gpr31
|
G protein-coupled receptor 31 |
| chr7_-_36000906 | 14.97 |
ENSRNOT00000011178
|
Plxnc1
|
plexin C1 |
| chr1_+_277190964 | 14.65 |
ENSRNOT00000080511
|
Casp7
|
caspase 7 |
| chr1_-_167093560 | 14.64 |
ENSRNOT00000027301
|
Il18bp
|
interleukin 18 binding protein |
| chr4_+_62380914 | 14.47 |
ENSRNOT00000029845
|
Tmem140
|
transmembrane protein 140 |
| chrX_+_11648989 | 14.36 |
ENSRNOT00000041003
|
Bcor
|
BCL6 co-repressor |
| chr5_-_56536772 | 14.22 |
ENSRNOT00000060765
|
Ddx58
|
DEXD/H-box helicase 58 |
| chr10_-_35175647 | 14.10 |
ENSRNOT00000078960
|
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr5_+_157423213 | 13.86 |
ENSRNOT00000023431
|
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr16_+_50111306 | 12.84 |
ENSRNOT00000019302
|
Cyp4v3
|
cytochrome P450, family 4, subfamily v, polypeptide 3 |
| chr4_-_120408232 | 12.78 |
ENSRNOT00000073269
|
LOC100911337
|
40S ribosomal protein S25-like |
| chr6_+_57396220 | 12.46 |
ENSRNOT00000044779
|
RGD1563613
|
similar to 40S ribosomal protein S25 |
| chr7_+_2691369 | 12.36 |
ENSRNOT00000049536
|
Stat2
|
signal transducer and activator of transcription 2 |
| chr6_+_8669722 | 11.91 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr8_+_128133431 | 11.72 |
ENSRNOT00000020174
|
Exog
|
exo/endonuclease G |
| chr3_-_163814799 | 11.72 |
ENSRNOT00000055127
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr1_+_247519939 | 10.92 |
ENSRNOT00000034421
|
Cd274
|
CD274 molecule |
| chr11_-_29638922 | 10.77 |
ENSRNOT00000048888
|
LOC100359671
|
ribosomal protein L29-like |
| chr2_+_196120580 | 10.36 |
ENSRNOT00000068528
|
Rfx5
|
regulatory factor X5 |
| chr10_-_87136026 | 10.21 |
ENSRNOT00000014230
ENSRNOT00000083233 |
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr10_-_16045835 | 10.21 |
ENSRNOT00000064832
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr4_-_88649216 | 9.82 |
ENSRNOT00000058626
|
Herc6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr12_-_41266430 | 9.70 |
ENSRNOT00000001853
ENSRNOT00000081108 |
Oas1b
|
2-5 oligoadenylate synthetase 1B |
| chr16_-_71203609 | 9.58 |
ENSRNOT00000088458
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr9_+_119517101 | 9.17 |
ENSRNOT00000020476
|
Lpin2
|
lipin 2 |
| chr14_+_19319299 | 9.16 |
ENSRNOT00000086542
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr15_-_93748742 | 8.87 |
ENSRNOT00000093370
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr13_-_102643223 | 8.75 |
ENSRNOT00000003155
|
Hlx
|
H2.0-like homeobox |
| chr3_-_2770620 | 8.57 |
ENSRNOT00000008253
|
Traf2
|
Tnf receptor-associated factor 2 |
| chr5_+_159735008 | 8.48 |
ENSRNOT00000064310
|
Rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr6_-_21135880 | 8.46 |
ENSRNOT00000051239
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chr4_-_120041238 | 8.46 |
ENSRNOT00000073799
|
LOC100911337
|
40S ribosomal protein S25-like |
| chr1_+_84009268 | 8.26 |
ENSRNOT00000057230
ENSRNOT00000081121 |
RGD1560854
|
similar to FLJ41131 protein |
| chr7_+_2635743 | 8.13 |
ENSRNOT00000004223
|
Mip
|
major intrinsic protein of lens fiber |
| chr4_-_120414118 | 8.05 |
ENSRNOT00000072795
|
LOC100911337
|
40S ribosomal protein S25-like |
| chr5_+_159734838 | 7.99 |
ENSRNOT00000079905
|
Rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr3_+_72460889 | 7.84 |
ENSRNOT00000012209
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr16_+_18690649 | 7.76 |
ENSRNOT00000015190
|
Mat1a
|
methionine adenosyltransferase 1A |
| chr14_-_81679082 | 7.36 |
ENSRNOT00000077799
|
Rnf4
|
ring finger protein 4 |
| chr2_+_1410934 | 6.98 |
ENSRNOT00000013625
ENSRNOT00000080222 |
Erap1
|
endoplasmic reticulum aminopeptidase 1 |
| chr5_-_24489468 | 6.91 |
ENSRNOT00000081395
|
Ints8
|
integrator complex subunit 8 |
| chr6_-_102196138 | 6.84 |
ENSRNOT00000014132
|
Tmem229b
|
transmembrane protein 229B |
| chr8_-_108261021 | 6.80 |
ENSRNOT00000040710
|
LOC100912210
|
40S ribosomal protein S25-like |
| chr1_+_53220397 | 5.80 |
ENSRNOT00000089989
|
AABR07001573.1
|
|
| chr1_+_64046377 | 5.49 |
ENSRNOT00000085010
|
Tmc4
|
transmembrane channel-like 4 |
| chr19_-_19377492 | 5.35 |
ENSRNOT00000080964
|
Nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr2_+_189062443 | 5.34 |
ENSRNOT00000028181
|
Adar
|
adenosine deaminase, RNA-specific |
| chr1_+_221043119 | 4.85 |
ENSRNOT00000028185
|
Map3k11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr1_+_72756809 | 4.78 |
ENSRNOT00000082423
|
Ppp6r1
|
protein phosphatase 6, regulatory subunit 1 |
| chr6_-_1466201 | 4.75 |
ENSRNOT00000089185
|
Eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr2_-_148722263 | 4.61 |
ENSRNOT00000017868
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr13_-_52514875 | 4.56 |
ENSRNOT00000064758
|
Nav1
|
neuron navigator 1 |
| chr1_+_64114721 | 4.48 |
ENSRNOT00000080466
|
Tmc4
|
transmembrane channel-like 4 |
| chr1_+_255564409 | 4.38 |
ENSRNOT00000081786
|
Btaf1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr17_-_84247038 | 4.18 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr5_-_78183122 | 4.07 |
ENSRNOT00000002083
ENSRNOT00000067076 |
Fkbp15
|
FK506 binding protein 15 |
| chr18_+_56364620 | 3.63 |
ENSRNOT00000068535
ENSRNOT00000086033 |
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr1_+_219439953 | 3.46 |
ENSRNOT00000025282
|
Ppp1ca
|
protein phosphatase 1 catalytic subunit alpha |
| chrX_+_128409472 | 3.31 |
ENSRNOT00000078482
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr7_-_101140308 | 3.25 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chr5_+_141572536 | 3.17 |
ENSRNOT00000023514
|
Rragc
|
Ras-related GTP binding C |
| chr12_-_9501213 | 3.14 |
ENSRNOT00000071942
|
Pdx1
|
pancreatic and duodenal homeobox 1 |
| chr1_-_102849430 | 3.13 |
ENSRNOT00000086856
|
Saa4
|
serum amyloid A4 |
| chr5_+_156603725 | 2.65 |
ENSRNOT00000075852
|
Hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr9_-_93125014 | 2.56 |
ENSRNOT00000023829
|
Htr2b
|
5-hydroxytryptamine receptor 2B |
| chr5_-_131860637 | 2.47 |
ENSRNOT00000064569
ENSRNOT00000080242 |
Slc5a9
|
solute carrier family 5 member 9 |
| chr1_+_41323194 | 1.65 |
ENSRNOT00000026350
|
Esr1
|
estrogen receptor 1 |
| chr8_-_62948720 | 1.54 |
ENSRNOT00000075476
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr20_+_8325041 | 1.50 |
ENSRNOT00000000640
|
Cmtr1
|
cap methyltransferase 1 |
| chr1_-_198008893 | 1.34 |
ENSRNOT00000025950
|
Il27
|
interleukin 27 |
| chr4_-_13565838 | 0.92 |
ENSRNOT00000048882
|
Olr987
|
olfactory receptor 987 |
| chr20_+_55253686 | 0.62 |
ENSRNOT00000057016
|
Ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr7_+_132857628 | 0.49 |
ENSRNOT00000005438
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr1_-_47213749 | 0.18 |
ENSRNOT00000024656
|
Dynlt1
|
dynein light chain Tctex-type 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 44.7 | 178.7 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 21.6 | 64.7 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 19.5 | 58.5 | GO:0046967 | cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 18.9 | 75.7 | GO:0050904 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) diapedesis(GO:0050904) cellular response to vitamin K(GO:0071307) |
| 16.1 | 64.4 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) cellular response to molecule of fungal origin(GO:0071226) |
| 16.1 | 48.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 15.3 | 45.8 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) |
| 14.6 | 73.0 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 14.5 | 72.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 13.1 | 39.4 | GO:0070212 | regulation of chromatin assembly(GO:0010847) protein poly-ADP-ribosylation(GO:0070212) |
| 13.0 | 117.1 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 13.0 | 39.0 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 12.2 | 133.7 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 11.9 | 35.6 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 11.2 | 44.8 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 11.0 | 132.2 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 10.8 | 43.1 | GO:1901423 | response to benzene(GO:1901423) |
| 10.7 | 64.1 | GO:0045345 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 8.5 | 34.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 8.5 | 25.4 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 7.9 | 15.9 | GO:0002481 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 7.9 | 23.7 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 7.7 | 46.3 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 7.7 | 23.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 7.3 | 36.6 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 7.0 | 55.8 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 7.0 | 27.8 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 6.9 | 13.8 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 6.9 | 27.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 6.5 | 45.6 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 6.4 | 19.2 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 6.4 | 32.0 | GO:0052041 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 6.3 | 101.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 6.3 | 25.3 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 6.3 | 176.2 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 6.2 | 49.9 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 5.7 | 45.5 | GO:0018377 | protein myristoylation(GO:0018377) |
| 5.5 | 38.8 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 5.3 | 16.0 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 5.3 | 21.1 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 3.9 | 31.2 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 3.7 | 36.9 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 3.6 | 14.4 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 3.6 | 14.2 | GO:0009597 | detection of virus(GO:0009597) |
| 3.5 | 28.3 | GO:0036018 | response to cobalamin(GO:0033590) cellular response to erythropoietin(GO:0036018) positive regulation of protein kinase C signaling(GO:0090037) |
| 3.3 | 65.7 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 3.2 | 16.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 3.2 | 19.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 3.0 | 107.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 3.0 | 26.6 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 2.9 | 14.6 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 2.9 | 40.9 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 2.9 | 8.6 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 2.8 | 14.1 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 2.8 | 22.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 2.8 | 30.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 2.7 | 30.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 2.7 | 35.4 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 2.7 | 5.3 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 2.2 | 8.7 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 2.2 | 34.5 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 2.1 | 12.4 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 1.9 | 19.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 1.9 | 25.0 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 1.8 | 7.4 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 1.7 | 40.4 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 1.6 | 116.6 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 1.6 | 34.4 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 1.6 | 14.6 | GO:1900115 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 1.4 | 195.5 | GO:0051607 | defense response to virus(GO:0051607) |
| 1.4 | 16.9 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 1.4 | 4.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.4 | 17.9 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 1.2 | 42.0 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 1.2 | 15.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 1.2 | 19.4 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 1.2 | 3.6 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 1.2 | 8.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 1.0 | 25.7 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 1.0 | 4.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.9 | 2.7 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.9 | 27.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.9 | 10.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.8 | 17.5 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) |
| 0.8 | 3.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.7 | 8.9 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.7 | 42.7 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.5 | 31.6 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.5 | 15.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.5 | 2.6 | GO:0071502 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) cellular response to temperature stimulus(GO:0071502) |
| 0.5 | 50.9 | GO:0031016 | pancreas development(GO:0031016) |
| 0.5 | 1.5 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.4 | 3.3 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.4 | 1.7 | GO:1990375 | baculum development(GO:1990375) |
| 0.4 | 26.4 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.4 | 16.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.4 | 4.4 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.3 | 4.8 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 7.8 | GO:0031954 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) positive regulation of protein autophosphorylation(GO:0031954) |
| 0.3 | 3.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 6.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.3 | 4.6 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.3 | 9.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.2 | 0.5 | GO:0035564 | regulation of kidney size(GO:0035564) |
| 0.2 | 0.2 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.2 | 21.5 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.2 | 52.4 | GO:0045087 | innate immune response(GO:0045087) |
| 0.1 | 12.8 | GO:0019395 | fatty acid oxidation(GO:0019395) |
| 0.1 | 53.4 | GO:0006935 | chemotaxis(GO:0006935) |
| 0.1 | 32.2 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.1 | 4.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 3.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 4.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 66.5 | GO:0098655 | cation transmembrane transport(GO:0098655) |
| 0.1 | 11.7 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 4.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 2.5 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 8.5 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.6 | 129.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 11.7 | 82.1 | GO:0042825 | TAP complex(GO:0042825) |
| 10.0 | 30.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 5.8 | 23.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 5.3 | 16.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 5.0 | 35.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 3.8 | 105.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 3.1 | 36.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 2.7 | 8.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 2.5 | 27.9 | GO:0042555 | MCM complex(GO:0042555) |
| 2.2 | 13.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.4 | 70.9 | GO:0016235 | aggresome(GO:0016235) |
| 1.4 | 27.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 1.2 | 21.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 1.1 | 46.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 1.1 | 19.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 1.1 | 5.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 1.0 | 8.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 1.0 | 323.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 1.0 | 24.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.9 | 22.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.9 | 10.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.6 | 15.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.5 | 8.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.5 | 23.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.5 | 36.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.5 | 46.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.4 | 6.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.4 | 26.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.3 | 3.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 3.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.3 | 18.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.3 | 16.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 206.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.3 | 26.8 | GO:0016605 | PML body(GO:0016605) |
| 0.3 | 337.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.3 | 22.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.2 | 26.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.2 | 14.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 4.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 107.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.2 | 90.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.2 | 521.7 | GO:0005829 | cytosol(GO:0005829) |
| 0.1 | 24.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 4.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 4.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 9.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 0.2 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 3.2 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 107.3 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 26.4 | 132.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 22.8 | 68.4 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 19.5 | 58.5 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 19.2 | 76.8 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 12.9 | 90.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 11.3 | 102.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 8.1 | 48.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 7.9 | 23.7 | GO:0046977 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 6.5 | 148.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 6.3 | 25.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 6.2 | 36.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 6.0 | 18.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 5.4 | 16.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 5.3 | 16.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 5.0 | 44.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 4.5 | 135.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 4.5 | 120.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 4.3 | 30.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 4.3 | 25.7 | GO:0048030 | disaccharide binding(GO:0048030) |
| 4.2 | 42.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 3.8 | 19.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 3.8 | 45.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 3.4 | 37.6 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 2.9 | 64.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 2.9 | 35.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 2.8 | 25.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 2.8 | 97.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 2.6 | 23.0 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 2.5 | 50.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 2.3 | 88.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 2.1 | 19.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.9 | 94.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 1.7 | 27.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 1.7 | 96.3 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 1.6 | 4.8 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 1.6 | 30.1 | GO:0001848 | complement binding(GO:0001848) |
| 1.4 | 9.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.3 | 28.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 1.3 | 99.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 1.3 | 14.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 1.3 | 384.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 1.2 | 65.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 1.2 | 7.4 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 1.2 | 35.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 1.2 | 4.8 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 1.2 | 3.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.1 | 7.8 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 1.1 | 15.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 1.1 | 11.7 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 1.0 | 45.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 1.0 | 53.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.9 | 10.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.9 | 86.3 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.8 | 14.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.8 | 39.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.8 | 5.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.8 | 62.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.7 | 37.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.7 | 28.6 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.6 | 8.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.6 | 41.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.6 | 25.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.5 | 9.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.5 | 1.5 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.4 | 1.7 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.3 | 97.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.2 | 2.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 4.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 11.9 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 34.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 3.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 27.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.2 | 66.5 | GO:0005261 | cation channel activity(GO:0005261) |
| 0.2 | 2.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.2 | 12.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 30.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 15.9 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.1 | 46.8 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.1 | 2.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 9.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 4.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 8.5 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.1 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 4.8 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 68.3 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 18.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.3 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 9.9 | GO:0008270 | zinc ion binding(GO:0008270) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.2 | 148.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 5.5 | 76.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 2.9 | 91.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 2.2 | 55.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 2.0 | 48.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 2.0 | 75.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 1.1 | 36.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 1.1 | 35.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 1.1 | 24.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 1.1 | 35.1 | PID MYC PATHWAY | C-MYC pathway |
| 1.1 | 36.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.9 | 40.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.8 | 18.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.8 | 25.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.8 | 36.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.8 | 63.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.7 | 36.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.7 | 10.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.7 | 23.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.6 | 190.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.6 | 95.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.5 | 14.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.4 | 1.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.4 | 8.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.4 | 5.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 16.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 4.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.3 | 3.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.3 | 11.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.3 | 62.0 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.3 | 27.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 5.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 3.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.1 | 665.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 7.1 | 77.8 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 6.1 | 304.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 5.5 | 71.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 5.3 | 15.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 4.7 | 225.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 4.5 | 126.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 4.3 | 8.6 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 4.1 | 28.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 3.9 | 65.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 2.5 | 27.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 2.4 | 16.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 2.4 | 35.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 2.3 | 30.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 2.2 | 22.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 1.9 | 55.9 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 1.7 | 64.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 1.6 | 23.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 1.4 | 30.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.4 | 64.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 1.3 | 12.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.9 | 15.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.9 | 21.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.8 | 19.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.7 | 8.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.7 | 9.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.6 | 14.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.4 | 5.3 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.3 | 56.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 3.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 8.5 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.2 | 2.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 4.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 4.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 13.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 3.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 4.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 3.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |