Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
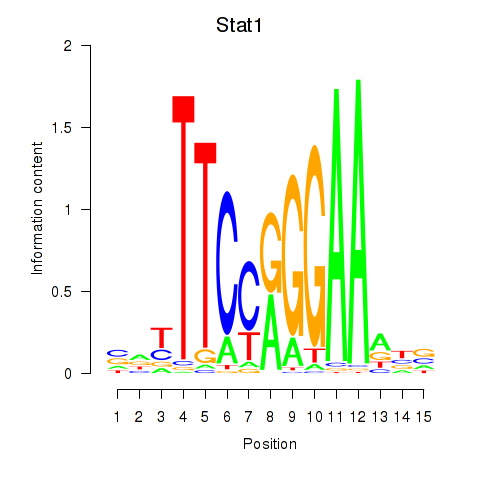
Results for Stat1
Z-value: 0.84
Transcription factors associated with Stat1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat1
|
ENSRNOG00000014079 | signal transducer and activator of transcription 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat1 | rn6_v1_chr9_-_54327958_54327958 | 0.61 | 2.3e-34 | Click! |
Activity profile of Stat1 motif
Sorted Z-values of Stat1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_23148396 | 40.95 |
ENSRNOT00000075237
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr1_-_98493978 | 40.89 |
ENSRNOT00000023942
|
Nkg7
|
natural killer cell granule protein 7 |
| chr15_-_34392066 | 36.74 |
ENSRNOT00000027315
|
Tgm1
|
transglutaminase 1 |
| chr3_-_72171078 | 36.48 |
ENSRNOT00000009817
|
Serping1
|
serpin family G member 1 |
| chr19_+_54314865 | 27.91 |
ENSRNOT00000024069
|
Irf8
|
interferon regulatory factor 8 |
| chr1_-_189181901 | 26.33 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr10_+_69412017 | 25.40 |
ENSRNOT00000009448
|
Ccl2
|
C-C motif chemokine ligand 2 |
| chr1_-_176079125 | 25.25 |
ENSRNOT00000047044
|
RGD1566189
|
similar to ferritin light chain |
| chr10_-_56506446 | 24.44 |
ENSRNOT00000021357
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr8_+_127735258 | 22.58 |
ENSRNOT00000015888
|
Vill
|
villin-like |
| chr15_-_35417273 | 21.59 |
ENSRNOT00000083961
ENSRNOT00000041430 |
Gzmb
|
granzyme B |
| chr14_+_17195014 | 21.41 |
ENSRNOT00000031667
|
Cxcl11
|
C-X-C motif chemokine ligand 11 |
| chr13_+_92103123 | 21.00 |
ENSRNOT00000004643
|
Olr1584
|
olfactory receptor 1584 |
| chr7_-_139318455 | 20.99 |
ENSRNOT00000092029
|
Hdac7
|
histone deacetylase 7 |
| chr9_+_10952374 | 20.57 |
ENSRNOT00000074993
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr14_-_23604834 | 20.28 |
ENSRNOT00000002760
|
Stap1
|
signal transducing adaptor family member 1 |
| chr10_+_39109522 | 18.83 |
ENSRNOT00000010968
|
Irf1
|
interferon regulatory factor 1 |
| chr3_-_160139947 | 18.79 |
ENSRNOT00000014151
|
Ada
|
adenosine deaminase |
| chr18_+_86299463 | 18.28 |
ENSRNOT00000058152
|
Cd226
|
CD226 molecule |
| chr15_-_36472327 | 17.84 |
ENSRNOT00000032601
ENSRNOT00000059660 |
LOC102553861
|
granzyme-like protein 1-like |
| chr20_-_3793985 | 17.35 |
ENSRNOT00000049540
ENSRNOT00000086293 |
RT1-CE16
|
RT1 class I, locus CE16 |
| chr1_+_154131926 | 17.25 |
ENSRNOT00000035257
|
LOC102549852
|
ferritin light chain 1-like |
| chr2_+_225827504 | 16.89 |
ENSRNOT00000018343
|
Gclm
|
glutamate cysteine ligase, modifier subunit |
| chr18_-_31430973 | 16.87 |
ENSRNOT00000026065
|
Pcdh12
|
protocadherin 12 |
| chr1_+_257157264 | 16.81 |
ENSRNOT00000067149
ENSRNOT00000079219 |
Plce1
|
phospholipase C, epsilon 1 |
| chr7_+_16404755 | 16.69 |
ENSRNOT00000044977
|
LOC108349290
|
olfactory receptor 6C70-like |
| chr18_-_36285444 | 16.64 |
ENSRNOT00000087852
|
Prelid2
|
PRELI domain containing 2 |
| chr1_-_78180216 | 16.09 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr2_+_199162745 | 16.08 |
ENSRNOT00000023488
|
Gja5
|
gap junction protein, alpha 5 |
| chr11_+_68105369 | 15.79 |
ENSRNOT00000046888
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr14_-_19072677 | 15.68 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr8_-_48850671 | 15.64 |
ENSRNOT00000016580
|
Cxcr5
|
C-X-C motif chemokine receptor 5 |
| chr20_-_3397039 | 15.60 |
ENSRNOT00000001084
ENSRNOT00000085259 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr15_+_34282936 | 15.04 |
ENSRNOT00000026364
|
Irf9
|
interferon regulatory factor 9 |
| chr1_-_213921208 | 14.18 |
ENSRNOT00000044393
|
Ano9
|
anoctamin 9 |
| chr2_+_30685840 | 14.16 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chr7_+_1206648 | 14.08 |
ENSRNOT00000073689
|
Pros1
|
protein S (alpha) |
| chr6_-_76652757 | 13.73 |
ENSRNOT00000042177
|
LOC100359668
|
ferritin light chain 2 |
| chr10_+_69476775 | 13.56 |
ENSRNOT00000031626
|
Ccl12
|
chemokine (C-C motif) ligand 12 |
| chr5_+_133896141 | 13.45 |
ENSRNOT00000011434
|
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr10_+_84167331 | 12.91 |
ENSRNOT00000010965
|
Hoxb4
|
homeo box B4 |
| chr8_+_116054465 | 12.70 |
ENSRNOT00000040056
|
Cish
|
cytokine inducible SH2-containing protein |
| chr17_-_15467320 | 12.45 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chr5_-_153625869 | 12.26 |
ENSRNOT00000024464
|
Clic4
|
chloride intracellular channel 4 |
| chrX_+_70461718 | 12.23 |
ENSRNOT00000078233
ENSRNOT00000003789 |
Kif4a
|
kinesin family member 4A |
| chr15_-_103927592 | 10.67 |
ENSRNOT00000031036
ENSRNOT00000013366 |
Abcc4
|
ATP binding cassette subfamily C member 4 |
| chrX_+_77065397 | 10.62 |
ENSRNOT00000090007
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr3_+_161519743 | 10.62 |
ENSRNOT00000055148
|
Cd40
|
CD40 molecule |
| chr19_+_15294248 | 10.50 |
ENSRNOT00000024622
|
Ces1f
|
carboxylesterase 1F |
| chr14_-_80973456 | 10.36 |
ENSRNOT00000013257
|
Hgfac
|
HGF activator |
| chr1_-_7480825 | 10.12 |
ENSRNOT00000048754
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr10_+_105393072 | 9.86 |
ENSRNOT00000013359
|
Ubald2
|
UBA-like domain containing 2 |
| chrX_-_25590048 | 9.67 |
ENSRNOT00000004873
|
Mid1
|
midline 1 |
| chrX_+_156854594 | 9.50 |
ENSRNOT00000083442
|
Renbp
|
renin binding protein |
| chr7_-_3342491 | 9.32 |
ENSRNOT00000081756
|
Rdh5
|
retinol dehydrogenase 5 |
| chr8_-_82492243 | 9.29 |
ENSRNOT00000013852
|
Tmod3
|
tropomodulin 3 |
| chr6_-_36940868 | 9.17 |
ENSRNOT00000006470
|
Gen1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr7_-_70328148 | 9.02 |
ENSRNOT00000074645
|
Mettl21b
|
methyltransferase like 21B |
| chr7_-_3074359 | 8.95 |
ENSRNOT00000019738
|
Ikzf4
|
IKAROS family zinc finger 4 |
| chr12_+_7081895 | 8.75 |
ENSRNOT00000047163
|
Hmgb1
|
high mobility group box 1 |
| chr2_+_53109684 | 8.67 |
ENSRNOT00000086590
|
Selenop
|
selenoprotein P |
| chr7_-_137856485 | 8.57 |
ENSRNOT00000007003
|
LOC688906
|
similar to splicing factor, arginine/serine-rich 2, interacting protein |
| chr3_-_52998650 | 8.52 |
ENSRNOT00000009988
|
RGD1560831
|
similar to 40S ribosomal protein S3 |
| chr14_+_81488008 | 8.32 |
ENSRNOT00000018477
|
Tnip2
|
TNFAIP3 interacting protein 2 |
| chr3_+_121632043 | 8.14 |
ENSRNOT00000024882
|
Polr1b
|
RNA polymerase I subunit B |
| chr8_+_133029625 | 8.14 |
ENSRNOT00000008809
|
Ccr3
|
C-C motif chemokine receptor 3 |
| chr11_+_80255790 | 8.14 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr2_+_165601007 | 8.12 |
ENSRNOT00000013931
|
Smc4
|
structural maintenance of chromosomes 4 |
| chr10_+_14240219 | 7.79 |
ENSRNOT00000020233
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr7_+_143707237 | 7.70 |
ENSRNOT00000074212
|
Tns2
|
tensin 2 |
| chr10_-_59883839 | 7.56 |
ENSRNOT00000093579
|
Aspa
|
aspartoacylase |
| chr11_+_31806618 | 7.14 |
ENSRNOT00000043410
ENSRNOT00000002769 |
Son
|
Son DNA binding protein |
| chr1_-_89042176 | 7.11 |
ENSRNOT00000080842
|
Kmt2b
|
lysine methyltransferase 2B |
| chr5_-_79874671 | 6.96 |
ENSRNOT00000084563
|
Tnc
|
tenascin C |
| chr8_-_116349896 | 6.80 |
ENSRNOT00000087306
|
Lsmem2
|
leucine-rich single-pass membrane protein 2 |
| chr7_-_145174771 | 6.61 |
ENSRNOT00000055270
|
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr17_+_9596957 | 6.52 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr4_+_123118468 | 6.35 |
ENSRNOT00000010895
|
Tmem43
|
transmembrane protein 43 |
| chr7_-_4293352 | 6.34 |
ENSRNOT00000047489
|
Olr984
|
olfactory receptor 984 |
| chr7_+_3900228 | 6.28 |
ENSRNOT00000071350
|
Olr921
|
olfactory receptor 921 |
| chr2_+_86996798 | 5.85 |
ENSRNOT00000070821
|
Zfp455
|
zinc finger protein 455 |
| chr7_-_102298522 | 5.75 |
ENSRNOT00000006273
|
A1bg
|
alpha-1-B glycoprotein |
| chr8_+_49621568 | 5.70 |
ENSRNOT00000021949
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr1_-_37818879 | 5.58 |
ENSRNOT00000043747
|
LOC680200
|
similar to zinc finger protein 455 |
| chr15_+_33004133 | 5.42 |
ENSRNOT00000013246
|
Oxa1l
|
OXA1L, mitochondrial inner membrane protein |
| chr10_-_95431312 | 5.38 |
ENSRNOT00000085552
|
AABR07030603.2
|
|
| chr10_-_89338739 | 5.25 |
ENSRNOT00000073923
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr4_-_69211671 | 5.25 |
ENSRNOT00000017852
|
LOC286960
|
preprotrypsinogen IV |
| chr7_+_6096690 | 5.20 |
ENSRNOT00000051694
|
Olr1006
|
olfactory receptor 1006 |
| chr2_-_140618405 | 5.17 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr10_-_65805693 | 5.13 |
ENSRNOT00000012245
|
Tnfaip1
|
TNF alpha induced protein 1 |
| chr9_+_42871950 | 4.96 |
ENSRNOT00000089673
|
Arid5a
|
AT-rich interaction domain 5A |
| chr2_+_211337271 | 4.79 |
ENSRNOT00000045155
|
Cox6b1
|
cytochrome c oxidase subunit 6B1 |
| chr17_+_57075218 | 4.57 |
ENSRNOT00000089536
|
Crem
|
cAMP responsive element modulator |
| chr1_-_134870255 | 4.50 |
ENSRNOT00000055829
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr20_+_4852671 | 4.50 |
ENSRNOT00000001111
|
Lta
|
lymphotoxin alpha |
| chr1_-_52894832 | 4.50 |
ENSRNOT00000016471
|
T
|
T brachyury transcription factor |
| chr3_-_161376119 | 4.37 |
ENSRNOT00000023521
|
Zfp335
|
zinc finger protein 335 |
| chr19_-_10884047 | 4.27 |
ENSRNOT00000088196
ENSRNOT00000063791 |
Cpne2
|
copine 2 |
| chr10_+_56605140 | 4.12 |
ENSRNOT00000024079
|
Phf23
|
PHD finger protein 23 |
| chr9_+_15166118 | 3.85 |
ENSRNOT00000020432
|
Mdfi
|
MyoD family inhibitor |
| chr1_+_144239020 | 3.73 |
ENSRNOT00000032106
|
Adamtsl3
|
ADAMTS-like 3 |
| chr15_+_34452116 | 3.71 |
ENSRNOT00000027647
|
Ltb4r
|
leukotriene B4 receptor |
| chr7_-_5818625 | 3.53 |
ENSRNOT00000051699
|
Olr886
|
olfactory receptor 886 |
| chr7_-_16432738 | 3.44 |
ENSRNOT00000049493
|
Olr932
|
olfactory receptor 932 |
| chr17_+_18358712 | 3.43 |
ENSRNOT00000001979
|
Nup153
|
nucleoporin 153 |
| chr6_-_42616548 | 3.43 |
ENSRNOT00000081433
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr12_-_23727535 | 3.42 |
ENSRNOT00000085911
ENSRNOT00000001950 |
Dtx2
|
deltex E3 ubiquitin ligase 2 |
| chr18_+_36829062 | 3.36 |
ENSRNOT00000025467
ENSRNOT00000086979 |
Tcerg1
|
transcription elongation regulator 1 |
| chr7_-_70355619 | 3.18 |
ENSRNOT00000031272
|
Tspan31
|
tetraspanin 31 |
| chr7_+_14037620 | 3.15 |
ENSRNOT00000009606
|
Syde1
|
synapse defective Rho GTPase homolog 1 |
| chrX_-_143453612 | 3.13 |
ENSRNOT00000051319
|
Atp11c
|
ATPase phospholipid transporting 11C |
| chr13_+_47739526 | 3.05 |
ENSRNOT00000006246
|
Il10
|
interleukin 10 |
| chr7_+_2718700 | 2.96 |
ENSRNOT00000050792
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr7_-_5168902 | 2.93 |
ENSRNOT00000090661
|
Olr894
|
olfactory receptor 894 |
| chr20_+_4852496 | 2.92 |
ENSRNOT00000088936
|
Lta
|
lymphotoxin alpha |
| chr4_+_109477920 | 2.87 |
ENSRNOT00000008468
|
Reg3a
|
regenerating islet-derived 3 alpha |
| chrX_-_70461553 | 2.84 |
ENSRNOT00000076110
ENSRNOT00000003872 ENSRNOT00000076812 |
Pdzd11
|
PDZ domain containing 11 |
| chr7_+_70328391 | 2.63 |
ENSRNOT00000075544
|
Mettl1
|
methyltransferase like 1 |
| chr14_-_42560174 | 2.59 |
ENSRNOT00000003128
|
Tmem33
|
transmembrane protein 33 |
| chr1_+_87790104 | 2.55 |
ENSRNOT00000074580
|
LOC103690114
|
zinc finger protein 383-like |
| chr13_+_90184569 | 2.52 |
ENSRNOT00000082393
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr3_+_77072410 | 2.47 |
ENSRNOT00000078540
|
Olr649
|
olfactory receptor 649 |
| chr3_-_74906989 | 2.44 |
ENSRNOT00000071001
|
Olr545
|
olfactory receptor 545 |
| chr2_-_211001258 | 2.43 |
ENSRNOT00000037336
|
Atxn7l2
|
ataxin 7-like 2 |
| chr2_+_86996497 | 2.41 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr10_+_10457320 | 2.40 |
ENSRNOT00000003832
|
Rbm25l1
|
RNA binding motif protein 25-like 1 |
| chr7_+_117390285 | 2.17 |
ENSRNOT00000016464
|
Exosc4
|
exosome component 4 |
| chr7_+_7461109 | 2.02 |
ENSRNOT00000074969
|
Olr1014
|
olfactory receptor 1014 |
| chr5_+_164951789 | 1.96 |
ENSRNOT00000055658
ENSRNOT00000012193 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr2_-_232117134 | 1.90 |
ENSRNOT00000030798
|
Alpk1
|
alpha-kinase 1 |
| chr7_+_5401508 | 1.85 |
ENSRNOT00000050898
|
Olr903
|
olfactory receptor 903 |
| chr1_-_164441167 | 1.83 |
ENSRNOT00000023935
|
Rps3
|
ribosomal protein S3 |
| chr11_-_82938357 | 1.73 |
ENSRNOT00000035945
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr9_-_116222374 | 1.71 |
ENSRNOT00000090111
ENSRNOT00000067900 |
Arhgap28
|
Rho GTPase activating protein 28 |
| chr19_-_25801526 | 1.62 |
ENSRNOT00000003884
|
Nacc1
|
nucleus accumbens associated 1 |
| chr2_+_182006242 | 1.58 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr5_+_101526551 | 1.57 |
ENSRNOT00000015254
|
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr1_-_167093560 | 1.56 |
ENSRNOT00000027301
|
Il18bp
|
interleukin 18 binding protein |
| chr14_-_86387606 | 1.53 |
ENSRNOT00000089384
ENSRNOT00000006642 |
Ddx56
|
DEAD-box helicase 56 |
| chr10_-_31052102 | 1.53 |
ENSRNOT00000007917
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like |
| chrX_+_159703578 | 1.51 |
ENSRNOT00000001162
|
Cd40lg
|
CD40 ligand |
| chr10_-_95262024 | 1.47 |
ENSRNOT00000066487
|
Bptf
|
bromodomain PHD finger transcription factor |
| chr7_-_3499697 | 1.46 |
ENSRNOT00000064311
|
Olr878
|
olfactory receptor 878 |
| chr10_+_17542374 | 1.41 |
ENSRNOT00000064079
|
Fbxw11
|
F-box and WD repeat domain containing 11 |
| chr1_+_157403595 | 1.31 |
ENSRNOT00000012781
|
Ccdc90b
|
coiled-coil domain containing 90B |
| chr7_-_5106708 | 1.16 |
ENSRNOT00000046001
|
Olr892
|
olfactory receptor 892 |
| chr13_-_45068077 | 1.03 |
ENSRNOT00000004969
|
Mcm6
|
minichromosome maintenance complex component 6 |
| chr7_+_11321057 | 0.90 |
ENSRNOT00000027737
|
Mrpl54
|
mitochondrial ribosomal protein L54 |
| chr5_+_165415136 | 0.89 |
ENSRNOT00000016317
ENSRNOT00000079407 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr2_+_189609800 | 0.78 |
ENSRNOT00000089016
|
Slc39a1
|
solute carrier family 39 member 1 |
| chr10_+_89339029 | 0.75 |
ENSRNOT00000031208
|
Rundc1
|
RUN domain containing 1 |
| chr5_-_151768123 | 0.54 |
ENSRNOT00000079380
|
Nudc
|
nuclear distribution C, dynein complex regulator |
| chr1_-_81412251 | 0.50 |
ENSRNOT00000026946
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr3_-_111873974 | 0.37 |
ENSRNOT00000077546
|
AABR07053509.2
|
|
| chr3_-_75168421 | 0.15 |
ENSRNOT00000072853
|
LOC103691842
|
olfactory receptor 10AG1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 25.4 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 7.2 | 21.6 | GO:0060545 | positive regulation of necroptotic process(GO:0060545) |
| 6.8 | 40.9 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 6.8 | 20.3 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 6.6 | 26.3 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 6.3 | 18.8 | GO:0070256 | germinal center B cell differentiation(GO:0002314) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) inosine biosynthetic process(GO:0046103) regulation of adenosine receptor signaling pathway(GO:0060167) negative regulation of mucus secretion(GO:0070256) |
| 6.1 | 18.3 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 5.4 | 16.1 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 5.4 | 16.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 4.7 | 14.2 | GO:2001226 | negative regulation of inorganic anion transmembrane transport(GO:1903796) negative regulation of chloride transport(GO:2001226) |
| 4.2 | 16.9 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 3.8 | 18.8 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 2.9 | 8.8 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) regulation of mismatch repair(GO:0032423) positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) T-helper 1 cell activation(GO:0035711) positive regulation of glycogen catabolic process(GO:0045819) negative regulation of apoptotic cell clearance(GO:2000426) |
| 2.7 | 37.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 2.6 | 39.6 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 2.6 | 10.5 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 2.4 | 9.5 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 2.3 | 7.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 2.1 | 21.0 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 2.0 | 8.1 | GO:0043380 | regulation of mast cell cytokine production(GO:0032763) regulation of memory T cell differentiation(GO:0043380) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 1.8 | 12.9 | GO:0048539 | bone marrow development(GO:0048539) |
| 1.8 | 5.4 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 1.7 | 12.2 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 1.7 | 12.1 | GO:0033590 | response to cobalamin(GO:0033590) |
| 1.6 | 8.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.4 | 21.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 1.4 | 4.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 1.3 | 21.4 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 1.3 | 27.9 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 1.3 | 15.0 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 1.2 | 10.8 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 1.2 | 9.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.1 | 9.2 | GO:0090267 | positive regulation of spindle checkpoint(GO:0090232) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 1.1 | 4.5 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) notochord formation(GO:0014028) |
| 1.0 | 15.6 | GO:0048535 | lymph node development(GO:0048535) |
| 0.9 | 12.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.9 | 16.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.9 | 6.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.9 | 8.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.9 | 9.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.9 | 4.4 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.8 | 20.6 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.7 | 15.7 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.7 | 10.7 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.7 | 2.6 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.6 | 2.6 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.6 | 3.7 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.6 | 1.8 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.5 | 5.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.5 | 8.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.5 | 3.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.5 | 2.0 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.5 | 2.8 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.4 | 7.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.4 | 3.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.4 | 7.6 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.4 | 2.2 | GO:0045006 | U4 snRNA 3'-end processing(GO:0034475) DNA deamination(GO:0045006) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.4 | 12.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.3 | 3.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.3 | 5.7 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.3 | 5.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.3 | 3.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.2 | 4.5 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.2 | 7.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.2 | 12.5 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.2 | 16.8 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.2 | 3.8 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.2 | 1.6 | GO:1900115 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.2 | 5.0 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.1 | 1.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.7 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 18.3 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 4.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.0 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 6.1 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.1 | 9.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 1.4 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.5 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 47.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 12.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 1.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 13.7 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 3.4 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 1.5 | GO:0001892 | embryonic placenta development(GO:0001892) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 25.4 | GO:0044299 | C-fiber(GO:0044299) |
| 2.7 | 21.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 2.7 | 18.8 | GO:0060205 | cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 2.7 | 10.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 1.3 | 7.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 1.0 | 8.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.9 | 10.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.8 | 16.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.7 | 3.4 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.7 | 8.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.6 | 10.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.6 | 3.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.5 | 64.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.5 | 2.6 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.4 | 2.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.4 | 7.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 51.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 6.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.3 | 3.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 21.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.3 | 19.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.3 | 26.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 12.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 12.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 1.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 2.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 11.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 1.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 9.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 8.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 4.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 10.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 3.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 5.4 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 15.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 95.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 5.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 4.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 11.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 9.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 7.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 3.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 3.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 6.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 9.0 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 4.7 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 9.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 62.5 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 6.5 | 39.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 5.4 | 16.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 4.3 | 21.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 4.0 | 16.1 | GO:0086075 | gap junction hemi-channel activity(GO:0055077) gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 3.7 | 36.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 2.5 | 7.6 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 2.2 | 40.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 2.1 | 37.4 | GO:0001848 | complement binding(GO:0001848) |
| 1.8 | 8.8 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 1.7 | 15.6 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 1.6 | 21.0 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 1.6 | 18.8 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 1.4 | 7.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 1.4 | 8.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 1.1 | 8.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.9 | 8.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.9 | 7.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.8 | 9.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.8 | 6.6 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.8 | 16.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.8 | 16.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.8 | 9.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.8 | 12.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.7 | 3.7 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.7 | 14.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.7 | 15.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.7 | 8.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.5 | 9.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 15.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.5 | 10.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.4 | 3.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.4 | 9.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.4 | 1.5 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.4 | 3.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.4 | 20.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.4 | 1.8 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.3 | 10.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 1.6 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.3 | 7.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.3 | 25.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.3 | 7.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 56.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 5.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 8.9 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 2.6 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.2 | 8.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 6.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 27.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 5.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 3.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 8.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 12.3 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 3.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 12.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 3.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 16.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 2.2 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.1 | 5.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 9.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 1.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 16.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 7.1 | GO:0043492 | ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.1 | 6.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 5.0 | GO:0044212 | regulatory region DNA binding(GO:0000975) transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 12.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 47.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 3.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 6.0 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 1.7 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.5 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 3.0 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 16.5 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 15.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 5.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 7.7 | GO:0019900 | kinase binding(GO:0019900) |
| 0.0 | 2.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.6 | GO:0008237 | metallopeptidase activity(GO:0008237) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 15.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 1.4 | 66.0 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.9 | 21.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.7 | 29.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.7 | 18.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.6 | 21.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.6 | 24.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.5 | 16.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.5 | 20.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.5 | 27.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.4 | 10.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.4 | 8.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 9.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.4 | 7.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.4 | 23.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 12.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 71.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 8.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.3 | 5.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 3.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 8.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 4.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 5.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 17.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 10.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 14.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 1.5 | 32.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.4 | 70.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 1.2 | 18.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 1.0 | 61.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.9 | 16.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.9 | 12.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.8 | 8.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.7 | 30.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.7 | 40.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.6 | 16.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.6 | 12.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.5 | 21.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.4 | 12.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.4 | 7.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 21.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.4 | 8.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.3 | 3.7 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 7.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 8.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 10.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 2.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.4 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.1 | 3.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 3.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 1.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 3.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 4.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.0 | REACTOME DNA REPAIR | Genes involved in DNA Repair |