Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
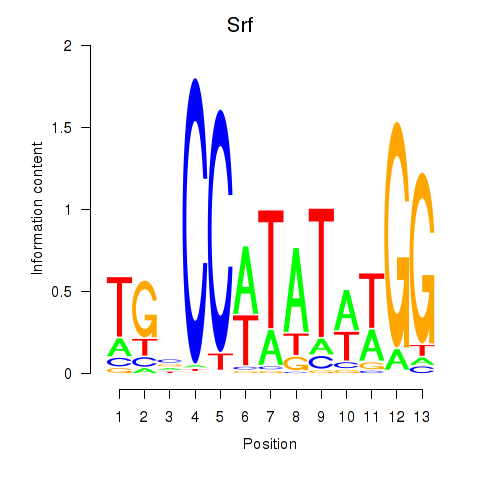
Results for Srf
Z-value: 2.37
Transcription factors associated with Srf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Srf
|
ENSRNOG00000018232 | serum response factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Srf | rn6_v1_chr9_+_16737642_16737642 | 0.71 | 7.1e-50 | Click! |
Activity profile of Srf motif
Sorted Z-values of Srf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_105512939 | 300.50 |
ENSRNOT00000011773
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr8_+_119030875 | 181.77 |
ENSRNOT00000028458
|
Myl3
|
myosin light chain 3 |
| chr6_-_127248372 | 161.82 |
ENSRNOT00000085517
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr5_+_164808323 | 159.99 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr10_+_70262361 | 151.56 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chrX_+_71342775 | 142.98 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr14_-_86146744 | 133.80 |
ENSRNOT00000019335
|
Myl7
|
myosin light chain 7 |
| chr10_+_53621375 | 120.01 |
ENSRNOT00000004147
|
Myh3
|
myosin heavy chain 3 |
| chr2_+_104416972 | 98.28 |
ENSRNOT00000017125
|
Trim55
|
tripartite motif-containing 55 |
| chr12_+_49761120 | 93.56 |
ENSRNOT00000070961
|
Myo18b
|
myosin XVIIIb |
| chr4_-_115239723 | 90.20 |
ENSRNOT00000042699
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr16_-_10941414 | 84.53 |
ENSRNOT00000086627
ENSRNOT00000085414 ENSRNOT00000081631 ENSRNOT00000087521 ENSRNOT00000083623 |
Ldb3
|
LIM domain binding 3 |
| chr1_-_25839198 | 83.03 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr4_-_99125111 | 79.50 |
ENSRNOT00000009184
|
Smyd1
|
SET and MYND domain containing 1 |
| chr1_-_215838209 | 78.47 |
ENSRNOT00000050760
|
Igf2
|
insulin-like growth factor 2 |
| chr16_+_49266903 | 74.64 |
ENSRNOT00000014704
|
Slc25a4
|
solute carrier family 25 member 4 |
| chr13_+_52662996 | 69.51 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr8_-_87282156 | 58.56 |
ENSRNOT00000087874
|
Filip1
|
filamin A interacting protein 1 |
| chr3_-_123607352 | 53.67 |
ENSRNOT00000051404
ENSRNOT00000028854 |
Adam33
|
ADAM metallopeptidase domain 33 |
| chr10_-_89338739 | 53.62 |
ENSRNOT00000073923
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr9_-_119332967 | 53.19 |
ENSRNOT00000021048
|
Myl12a
|
myosin light chain 12A |
| chr15_-_3544685 | 49.90 |
ENSRNOT00000015179
ENSRNOT00000085126 |
Vcl
|
vinculin |
| chr3_+_152857592 | 46.48 |
ENSRNOT00000027445
|
Myl9
|
myosin light chain 9 |
| chr1_-_198232344 | 41.69 |
ENSRNOT00000080988
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr10_-_62699723 | 40.51 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr7_-_117288018 | 39.41 |
ENSRNOT00000091285
|
Plec
|
plectin |
| chr2_-_198002625 | 36.95 |
ENSRNOT00000091888
|
BC028528
|
cDNA sequence BC028528 |
| chr1_+_199664173 | 33.84 |
ENSRNOT00000054980
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr7_+_12782491 | 31.43 |
ENSRNOT00000065093
|
Cnn2
|
calponin 2 |
| chr1_-_209641123 | 30.51 |
ENSRNOT00000021702
|
Ebf3
|
early B-cell factor 3 |
| chr15_+_32763067 | 29.65 |
ENSRNOT00000011998
|
AABR07017902.1
|
|
| chr9_+_16737642 | 28.87 |
ENSRNOT00000061430
|
Srf
|
serum response factor |
| chr10_+_18911938 | 28.53 |
ENSRNOT00000007408
|
Kcnmb1
|
potassium calcium-activated channel subfamily M regulatory beta subunit 1 |
| chr15_-_19876389 | 26.98 |
ENSRNOT00000012400
ENSRNOT00000086368 |
Fermt2
|
fermitin family member 2 |
| chr4_-_166803127 | 26.68 |
ENSRNOT00000067909
|
RGD1306750
|
LOC362451 |
| chr6_+_109300433 | 26.12 |
ENSRNOT00000010712
|
Fos
|
FBJ osteosarcoma oncogene |
| chr2_+_189423559 | 24.56 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
| chr9_+_45672157 | 24.47 |
ENSRNOT00000017882
|
Pdcl3
|
phosducin-like 3 |
| chr13_-_91872954 | 22.61 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr3_-_46726946 | 22.28 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr2_+_208749996 | 22.17 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr10_+_36098051 | 21.38 |
ENSRNOT00000083971
|
Adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr2_+_208750356 | 20.97 |
ENSRNOT00000041562
|
Chia
|
chitinase, acidic |
| chr1_+_46835971 | 19.31 |
ENSRNOT00000066798
|
Synj2
|
synaptojanin 2 |
| chr1_-_175796040 | 18.66 |
ENSRNOT00000024060
|
Mrvi1
|
murine retrovirus integration site 1 homolog |
| chr10_-_88060561 | 18.55 |
ENSRNOT00000019133
|
Krt19
|
keratin 19 |
| chr1_+_40816107 | 17.33 |
ENSRNOT00000060767
|
Akap12
|
A-kinase anchoring protein 12 |
| chr14_-_7384876 | 16.26 |
ENSRNOT00000086694
|
Aff1
|
AF4/FMR2 family, member 1 |
| chr10_+_13915214 | 15.82 |
ENSRNOT00000015092
|
Pkd1
|
polycystin 1, transient receptor potential channel interacting |
| chr15_-_2648551 | 15.77 |
ENSRNOT00000086864
|
Vdac2
|
voltage-dependent anion channel 2 |
| chr3_+_80555196 | 15.70 |
ENSRNOT00000067318
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr17_-_80577027 | 15.35 |
ENSRNOT00000057827
|
Rsu1
|
Ras suppressor protein 1 |
| chr1_+_220826560 | 15.31 |
ENSRNOT00000027891
|
Fosl1
|
FOS like 1, AP-1 transcription factor subunit |
| chr7_-_54778848 | 14.36 |
ENSRNOT00000005399
|
Glipr1
|
GLI pathogenesis-related 1 |
| chr6_+_76745981 | 13.30 |
ENSRNOT00000076784
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr5_+_139385429 | 11.29 |
ENSRNOT00000078622
|
Scmh1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr15_-_34693034 | 10.51 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr1_-_80221710 | 10.25 |
ENSRNOT00000091687
|
Fosb
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr4_-_145329878 | 9.52 |
ENSRNOT00000011827
|
Tada3
|
transcriptional adaptor 3 |
| chr3_-_161299024 | 9.47 |
ENSRNOT00000021216
|
Neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr16_-_3765917 | 9.21 |
ENSRNOT00000088284
|
Duxbl1
|
double homeobox B-like 1 |
| chr1_-_47502952 | 8.59 |
ENSRNOT00000025580
|
Tagap
|
T-cell activation RhoGTPase activating protein |
| chr2_+_35977828 | 8.45 |
ENSRNOT00000076712
|
LOC108348103
|
serine protease inhibitor Kazal-type 5-like |
| chr2_-_45077219 | 8.24 |
ENSRNOT00000014319
|
Gzmk
|
granzyme K |
| chrX_-_156440461 | 8.03 |
ENSRNOT00000083951
|
Rpl10
|
ribosomal protein L10 |
| chr13_-_36022197 | 7.94 |
ENSRNOT00000091280
|
Cfap221
|
cilia and flagella associated protein 221 |
| chr10_-_13814304 | 7.93 |
ENSRNOT00000012203
|
Dnase1l2
|
deoxyribonuclease 1 like 2 |
| chr15_-_54906203 | 7.90 |
ENSRNOT00000020186
|
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr18_-_38329721 | 7.23 |
ENSRNOT00000080220
ENSRNOT00000060125 |
RGD1563060
|
similar to AVLV472 |
| chr8_-_104593625 | 6.82 |
ENSRNOT00000016625
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr3_+_138024508 | 6.70 |
ENSRNOT00000007794
|
Dstn
|
destrin, actin depolymerizing factor |
| chr3_-_167759273 | 6.25 |
ENSRNOT00000049457
|
AABR07054721.1
|
|
| chr1_-_31262466 | 6.08 |
ENSRNOT00000029531
|
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chrX_-_102510007 | 5.71 |
ENSRNOT00000082854
|
AABR07040494.1
|
|
| chr1_-_193700673 | 5.08 |
ENSRNOT00000071314
|
Paip2l1
|
polyadenylate-binding protein-interacting protein 2-like 1 |
| chr19_-_11057254 | 3.83 |
ENSRNOT00000025559
|
Herpud1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr10_-_44026578 | 3.72 |
ENSRNOT00000050448
|
Olr1418
|
olfactory receptor 1418 |
| chr1_-_189666440 | 3.58 |
ENSRNOT00000019167
|
Dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr1_+_79474809 | 3.32 |
ENSRNOT00000044927
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr1_-_72339395 | 3.24 |
ENSRNOT00000021772
|
Zfp580
|
zinc finger protein 580 |
| chr13_+_80125391 | 3.23 |
ENSRNOT00000044190
|
Mir199a2
|
microRNA 199a-2 |
| chr8_+_60709851 | 2.59 |
ENSRNOT00000021817
|
Rcn2
|
reticulocalbin 2 |
| chr15_+_31642169 | 2.59 |
ENSRNOT00000072362
|
AABR07017833.1
|
|
| chr8_+_48569328 | 2.49 |
ENSRNOT00000084030
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr11_-_89510871 | 2.37 |
ENSRNOT00000035247
|
Prkdc
|
protein kinase, DNA activated, catalytic polypeptide |
| chr8_+_117780891 | 2.33 |
ENSRNOT00000077236
|
Shisa5
|
shisa family member 5 |
| chr1_+_87650207 | 2.14 |
ENSRNOT00000077161
|
Zfp84
|
zinc finger protein 84 |
| chr4_-_106983279 | 2.10 |
ENSRNOT00000007700
|
AABR07061152.1
|
|
| chr15_-_36417868 | 1.82 |
ENSRNOT00000028062
|
AABR07018028.1
|
|
| chr15_+_32012543 | 1.70 |
ENSRNOT00000078174
|
AABR07017868.5
|
|
| chr6_-_102047758 | 1.63 |
ENSRNOT00000012101
|
Atp6v1d
|
ATPase H+ transporting V1 subunit D |
| chr9_+_84689150 | 1.39 |
ENSRNOT00000072242
|
Kcne4
|
potassium voltage-gated channel subfamily E regulatory subunit 4 |
| chr11_+_31539016 | 1.20 |
ENSRNOT00000072856
|
Ifnar2
|
interferon alpha and beta receptor subunit 2 |
| chr3_+_159569363 | 1.17 |
ENSRNOT00000064159
|
Tox2
|
TOX high mobility group box family member 2 |
| chr1_-_212991658 | 1.10 |
ENSRNOT00000091487
|
Olr298
|
olfactory receptor 298 |
| chr6_+_94636222 | 1.07 |
ENSRNOT00000005846
|
Daam1
|
dishevelled associated activator of morphogenesis 1 |
| chr9_+_119873170 | 0.20 |
ENSRNOT00000037587
|
Mettl4
|
methyltransferase like 4 |
| chr4_-_176606382 | 0.18 |
ENSRNOT00000065576
|
Recql
|
RecQ like helicase |
| chr15_-_29443454 | 0.09 |
ENSRNOT00000082167
|
AABR07017635.2
|
|
| chr8_+_41336340 | 0.01 |
ENSRNOT00000072049
|
Olr1225
|
olfactory receptor 1225 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Srf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 78.1 | 390.7 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 53.3 | 160.0 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 27.7 | 83.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 23.2 | 69.5 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 18.7 | 74.6 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 11.2 | 78.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 9.6 | 28.9 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 8.6 | 43.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 7.8 | 93.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 7.1 | 28.5 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 5.3 | 15.8 | GO:0072237 | distal tubule morphogenesis(GO:0072156) metanephric proximal tubule development(GO:0072237) |
| 5.2 | 181.8 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 4.8 | 33.8 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 4.5 | 22.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 3.8 | 15.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 3.4 | 6.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 3.3 | 26.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 3.2 | 41.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 3.1 | 15.7 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 3.1 | 24.5 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 2.8 | 49.9 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 2.7 | 79.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 2.6 | 112.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 2.6 | 120.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 2.2 | 151.6 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 2.1 | 161.8 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 2.1 | 39.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 2.0 | 99.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 1.9 | 18.7 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 1.7 | 184.7 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 1.7 | 17.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 1.5 | 31.4 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 1.4 | 19.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 1.3 | 7.9 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 1.0 | 9.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.9 | 9.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.8 | 3.8 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.6 | 2.4 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.6 | 7.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 15.8 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.3 | 8.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 15.4 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.3 | 7.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.3 | 2.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 10.2 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.2 | 3.6 | GO:0010225 | response to UV-C(GO:0010225) protein neddylation(GO:0045116) |
| 0.2 | 13.3 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 8.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 24.6 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 11.3 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 3.2 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.1 | 2.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 1.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 6.8 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 1.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 27.7 | 83.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 20.2 | 161.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 17.4 | 69.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 14.0 | 210.2 | GO:0032982 | myosin filament(GO:0032982) |
| 11.1 | 99.7 | GO:0016460 | myosin II complex(GO:0016460) |
| 10.4 | 93.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 9.8 | 68.4 | GO:1990357 | terminal web(GO:1990357) |
| 7.4 | 357.3 | GO:0031672 | A band(GO:0031672) |
| 5.7 | 160.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 5.2 | 26.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 4.4 | 374.5 | GO:0042641 | actomyosin(GO:0042641) |
| 3.9 | 19.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 2.4 | 9.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 2.0 | 39.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 2.0 | 15.7 | GO:0097443 | sorting endosome(GO:0097443) |
| 1.9 | 227.5 | GO:0030018 | Z disc(GO:0030018) |
| 1.3 | 3.8 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.8 | 22.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.7 | 9.5 | GO:0030914 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.7 | 13.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.6 | 2.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.5 | 90.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.5 | 11.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 15.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.3 | 16.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.3 | 33.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.3 | 28.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 8.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 15.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 6.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 6.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 24.3 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 8.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 30.0 | GO:0005924 | cell-substrate adherens junction(GO:0005924) cell-substrate junction(GO:0030055) |
| 0.1 | 25.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 6.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.6 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.1 | 21.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 32.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 11.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 3.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.4 | 69.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 8.3 | 74.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 7.2 | 43.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 6.8 | 33.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 6.4 | 160.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 6.1 | 181.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 6.0 | 41.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 5.8 | 46.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 5.3 | 84.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 5.0 | 110.0 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 4.6 | 300.5 | GO:0017022 | myosin binding(GO:0017022) |
| 4.1 | 78.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 3.8 | 49.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 3.8 | 151.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 3.1 | 24.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 2.9 | 28.9 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 2.4 | 19.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 2.4 | 82.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 1.6 | 7.9 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 1.4 | 165.3 | GO:0005178 | integrin binding(GO:0005178) |
| 1.0 | 17.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.0 | 28.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 1.0 | 26.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.8 | 90.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.7 | 53.2 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.7 | 15.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.7 | 161.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.7 | 13.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.6 | 75.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.6 | 2.4 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.6 | 27.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.5 | 75.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.5 | 9.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.4 | 79.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.4 | 1.2 | GO:0019961 | interferon binding(GO:0019961) |
| 0.4 | 6.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.3 | 35.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.3 | 39.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.3 | 3.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 73.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 7.9 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.1 | 33.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 2.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 23.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 0.2 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 14.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 70.0 | GO:0005524 | ATP binding(GO:0005524) |
| 0.0 | 1.6 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 1.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 17.9 | GO:0042802 | identical protein binding(GO:0042802) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 55.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 1.6 | 79.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 1.6 | 134.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 1.3 | 49.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 1.2 | 22.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 1.1 | 78.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 1.1 | 46.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.6 | 15.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.5 | 36.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.5 | 18.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 54.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 12.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 7.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.2 | 395.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 9.3 | 186.6 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 5.7 | 160.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 3.7 | 78.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 2.5 | 42.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 2.2 | 74.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 1.9 | 47.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 1.6 | 26.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 1.4 | 39.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.1 | 41.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.6 | 148.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.6 | 7.9 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.5 | 19.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.4 | 21.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 22.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 24.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 3.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 14.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 8.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 1.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |