Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
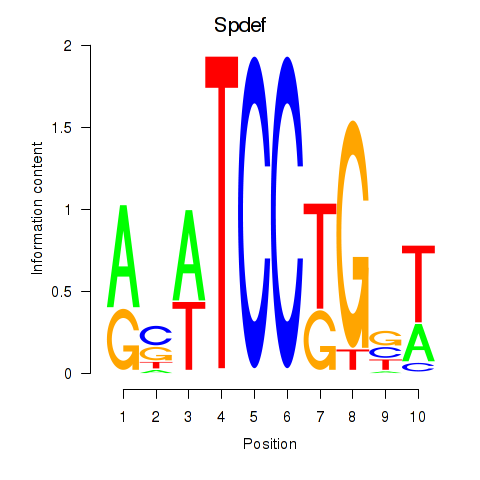
Results for Spdef
Z-value: 0.58
Transcription factors associated with Spdef
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Spdef
|
ENSRNOG00000000491 | SAM pointed domain containing ets transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Spdef | rn6_v1_chr20_-_7307280_7307284 | -0.41 | 2.6e-14 | Click! |
Activity profile of Spdef motif
Sorted Z-values of Spdef motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_89774764 | 20.23 |
ENSRNOT00000005619
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr13_-_55173692 | 18.48 |
ENSRNOT00000064785
ENSRNOT00000029878 ENSRNOT00000029865 ENSRNOT00000060292 ENSRNOT00000000814 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr7_+_143629455 | 15.09 |
ENSRNOT00000073951
|
Krt18
|
keratin 18 |
| chrX_+_128897181 | 13.96 |
ENSRNOT00000008444
|
Sh2d1a
|
SH2 domain containing 1A |
| chr10_+_11810926 | 13.24 |
ENSRNOT00000036205
ENSRNOT00000036189 |
Nlrc3
|
NLR family, CARD domain containing 3 |
| chr10_+_47930633 | 12.60 |
ENSRNOT00000003515
|
Grap
|
GRB2-related adaptor protein |
| chrX_+_134979646 | 12.52 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chrX_+_15155230 | 12.33 |
ENSRNOT00000073289
ENSRNOT00000051439 |
Was
|
Wiskott-Aldrich syndrome |
| chr8_+_55603968 | 12.19 |
ENSRNOT00000066848
|
Pou2af1
|
POU class 2 associating factor 1 |
| chr11_+_84745904 | 12.02 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr4_+_69138525 | 11.70 |
ENSRNOT00000073589
|
Trbv1
|
T cell receptor beta, variable 1 |
| chr18_+_44716226 | 11.13 |
ENSRNOT00000086431
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr14_-_64476796 | 10.86 |
ENSRNOT00000029104
|
Gba3
|
glucosidase, beta, acid 3 |
| chr1_+_260289589 | 10.74 |
ENSRNOT00000051058
|
Dntt
|
DNA nucleotidylexotransferase |
| chr20_+_5040337 | 10.51 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr4_-_163227242 | 9.83 |
ENSRNOT00000091552
ENSRNOT00000077793 |
Clec7a
|
C-type lectin domain family 7, member A |
| chr20_+_9948908 | 9.41 |
ENSRNOT00000001541
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr3_+_161018511 | 9.08 |
ENSRNOT00000019804
ENSRNOT00000039664 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr1_-_260254600 | 8.74 |
ENSRNOT00000019014
|
Blnk
|
B-cell linker |
| chr5_+_154522119 | 8.71 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr2_-_157759819 | 8.42 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr14_+_45062662 | 8.31 |
ENSRNOT00000059124
|
Tlr10
|
toll-like receptor 10 |
| chr10_-_104628676 | 8.21 |
ENSRNOT00000010466
|
Unc13d
|
unc-13 homolog D |
| chr7_+_11077411 | 8.14 |
ENSRNOT00000007117
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr1_+_201620642 | 7.82 |
ENSRNOT00000093674
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr15_-_33766438 | 7.71 |
ENSRNOT00000033977
|
Ap1g2
|
adaptor-related protein complex 1, gamma 2 subunit |
| chrX_+_115351114 | 7.44 |
ENSRNOT00000047553
|
Hmg1l1
|
high-mobility group (nonhistone chromosomal) protein 1-like 1 |
| chr5_-_169630340 | 7.44 |
ENSRNOT00000087043
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr1_-_85317968 | 7.41 |
ENSRNOT00000026891
|
Gmfg
|
glia maturation factor, gamma |
| chr13_+_52645257 | 7.38 |
ENSRNOT00000012801
|
Lad1
|
ladinin 1 |
| chr9_-_92616165 | 7.26 |
ENSRNOT00000056995
|
Sp110
|
SP110 nuclear body protein |
| chr9_+_94745217 | 7.18 |
ENSRNOT00000051338
|
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr2_+_183674522 | 7.03 |
ENSRNOT00000014433
|
Tmem154
|
transmembrane protein 154 |
| chr8_+_116054465 | 6.80 |
ENSRNOT00000040056
|
Cish
|
cytokine inducible SH2-containing protein |
| chr8_+_102304095 | 6.77 |
ENSRNOT00000011358
|
Slc9a9
|
solute carrier family 9 member A9 |
| chr10_-_10767389 | 6.73 |
ENSRNOT00000066754
|
Smim22
|
small integral membrane protein 22 |
| chr6_+_12253788 | 6.66 |
ENSRNOT00000061675
|
Ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr9_+_65620658 | 6.63 |
ENSRNOT00000084498
|
Casp8
|
caspase 8 |
| chr7_-_132984110 | 6.51 |
ENSRNOT00000046626
|
Hmgb1-ps3
|
high mobility group box 1, pseudogene 3 |
| chr3_+_148150698 | 6.38 |
ENSRNOT00000087075
ENSRNOT00000010251 |
Hm13
|
histocompatibility minor 13 |
| chr5_-_154319629 | 5.95 |
ENSRNOT00000014415
|
Lypla2
|
lysophospholipase II |
| chr4_+_92431710 | 5.92 |
ENSRNOT00000049438
|
LOC108350839
|
high mobility group protein B1-like |
| chr8_+_71914867 | 5.90 |
ENSRNOT00000023372
|
Dapk2
|
death-associated protein kinase 2 |
| chr10_+_84986328 | 5.90 |
ENSRNOT00000085830
ENSRNOT00000067665 |
Osbpl7
|
oxysterol binding protein-like 7 |
| chr1_+_199217016 | 5.87 |
ENSRNOT00000025732
|
Orai3
|
ORAI calcium release-activated calcium modulator 3 |
| chr12_+_2180150 | 5.64 |
ENSRNOT00000001322
|
Stxbp2
|
syntaxin binding protein 2 |
| chr13_+_5480150 | 5.61 |
ENSRNOT00000057873
|
AABR07019870.1
|
|
| chr1_+_84044551 | 5.56 |
ENSRNOT00000028290
|
Coq8b
|
coenzyme Q8B |
| chr1_-_43638161 | 5.50 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr1_-_146289465 | 5.47 |
ENSRNOT00000017362
|
Abhd17c
|
abhydrolase domain containing 17C |
| chr10_+_56576428 | 5.44 |
ENSRNOT00000079237
ENSRNOT00000023291 |
Cldn7
|
claudin 7 |
| chrX_-_72370044 | 5.43 |
ENSRNOT00000004224
|
Hdac8
|
histone deacetylase 8 |
| chr5_-_150704117 | 5.43 |
ENSRNOT00000067455
|
Sesn2
|
sestrin 2 |
| chr17_-_78812111 | 5.39 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr10_+_43744882 | 5.36 |
ENSRNOT00000078662
|
Zfp692
|
zinc finger protein 692 |
| chr16_-_19376776 | 5.25 |
ENSRNOT00000020748
|
Rab8a
|
RAB8A, member RAS oncogene family |
| chr4_-_60548021 | 5.09 |
ENSRNOT00000065546
|
AABR07060165.1
|
|
| chr4_+_170149029 | 5.04 |
ENSRNOT00000073287
|
LOC103690002
|
histone H2A.J |
| chr16_+_26483463 | 4.97 |
ENSRNOT00000079010
|
LOC100910404
|
actin, cytoplasmic 2-like |
| chr10_-_40657861 | 4.92 |
ENSRNOT00000058665
|
Fat2
|
FAT atypical cadherin 2 |
| chr10_-_57275708 | 4.87 |
ENSRNOT00000005370
|
Pfn1
|
profilin 1 |
| chr4_-_118595580 | 4.79 |
ENSRNOT00000024436
|
Anxa4
|
annexin A4 |
| chr14_-_62646110 | 4.77 |
ENSRNOT00000002992
|
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr19_+_37260469 | 4.76 |
ENSRNOT00000021362
|
Elmo3
|
engulfment and cell motility 3 |
| chr1_+_13595295 | 4.71 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr3_+_45277348 | 4.46 |
ENSRNOT00000059246
|
Pkp4
|
plakophilin 4 |
| chr3_-_120106697 | 4.44 |
ENSRNOT00000020354
|
Prom2
|
prominin 2 |
| chr3_-_9807328 | 4.44 |
ENSRNOT00000064660
|
Tor1a
|
torsin family 1, member A |
| chr3_+_122114108 | 4.41 |
ENSRNOT00000091935
|
Sirpa
|
signal-regulatory protein alpha |
| chr17_-_44520240 | 4.23 |
ENSRNOT00000086538
|
Hist1h2bh
|
histone cluster 1 H2B family member h |
| chr11_+_82680253 | 4.18 |
ENSRNOT00000077119
ENSRNOT00000075512 |
Liph
|
lipase H |
| chr6_+_126170911 | 4.17 |
ENSRNOT00000077477
|
Rin3
|
Ras and Rab interactor 3 |
| chr6_+_126170720 | 4.15 |
ENSRNOT00000065246
|
Rin3
|
Ras and Rab interactor 3 |
| chr10_-_15211325 | 4.04 |
ENSRNOT00000027083
|
Rhot2
|
ras homolog family member T2 |
| chr8_+_49378644 | 3.96 |
ENSRNOT00000007588
|
Jaml
|
junction adhesion molecule like |
| chr1_+_85386470 | 3.91 |
ENSRNOT00000093332
ENSRNOT00000044326 |
Plekhg2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr3_+_113131327 | 3.83 |
ENSRNOT00000018460
|
Tubgcp4
|
tubulin, gamma complex associated protein 4 |
| chr2_+_124400679 | 3.82 |
ENSRNOT00000058495
ENSRNOT00000067034 |
Spry1
|
sprouty RTK signaling antagonist 1 |
| chr8_+_133210473 | 3.81 |
ENSRNOT00000082774
|
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr2_-_210873024 | 3.76 |
ENSRNOT00000026051
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chrX_-_1741701 | 3.75 |
ENSRNOT00000031115
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr12_-_22021851 | 3.72 |
ENSRNOT00000039280
|
Tsc22d4
|
TSC22 domain family, member 4 |
| chr1_-_84168044 | 3.71 |
ENSRNOT00000084586
ENSRNOT00000028348 |
Shkbp1
|
Sh3kbp1 binding protein 1 |
| chr17_+_44520537 | 3.70 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr3_+_66673246 | 3.67 |
ENSRNOT00000081338
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr5_+_122647281 | 3.66 |
ENSRNOT00000066041
|
Mier1
|
mesoderm induction early response 1, transcriptional regulator |
| chr4_+_140247313 | 3.59 |
ENSRNOT00000040255
ENSRNOT00000064025 ENSRNOT00000041130 ENSRNOT00000043646 |
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr19_+_43392085 | 3.57 |
ENSRNOT00000023854
|
Sf3b3
|
splicing factor 3b, subunit 3 |
| chr3_+_164665532 | 3.51 |
ENSRNOT00000014309
|
Ptpn1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr7_+_2458264 | 3.50 |
ENSRNOT00000003550
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr2_+_225827504 | 3.46 |
ENSRNOT00000018343
|
Gclm
|
glutamate cysteine ligase, modifier subunit |
| chr11_-_33003021 | 3.45 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr17_-_21739408 | 3.45 |
ENSRNOT00000060335
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr2_+_196013799 | 3.35 |
ENSRNOT00000084023
|
Pogz
|
pogo transposable element with ZNF domain |
| chr18_-_24397551 | 3.34 |
ENSRNOT00000023032
|
Slc25a46
|
solute carrier family 25, member 46 |
| chr10_-_65437143 | 3.31 |
ENSRNOT00000017264
|
Nek8
|
NIMA-related kinase 8 |
| chr5_+_35991068 | 3.30 |
ENSRNOT00000061139
|
Pnisr
|
PNN interacting serine and arginine rich protein |
| chr1_-_134867001 | 3.29 |
ENSRNOT00000090149
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chrX_-_70461553 | 3.24 |
ENSRNOT00000076110
ENSRNOT00000003872 ENSRNOT00000076812 |
Pdzd11
|
PDZ domain containing 11 |
| chr14_+_77079402 | 3.23 |
ENSRNOT00000042200
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr3_+_2462466 | 3.21 |
ENSRNOT00000014087
|
Rnf208
|
ring finger protein 208 |
| chr18_-_69671199 | 3.19 |
ENSRNOT00000082484
|
Smad4
|
SMAD family member 4 |
| chr16_-_2367253 | 3.19 |
ENSRNOT00000083573
|
Pde12
|
phosphodiesterase 12 |
| chr10_-_97647822 | 3.16 |
ENSRNOT00000065781
|
AC106648.1
|
|
| chr15_+_34352914 | 3.15 |
ENSRNOT00000067150
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr14_-_106393670 | 3.13 |
ENSRNOT00000011429
|
Mdh1
|
malate dehydrogenase 1 |
| chr19_+_44164935 | 3.12 |
ENSRNOT00000048998
|
Gabarapl2
|
GABA type A receptor associated protein like 2 |
| chr4_+_66091641 | 3.11 |
ENSRNOT00000043147
|
AABR07060287.1
|
|
| chrX_-_71127237 | 3.08 |
ENSRNOT00000076403
ENSRNOT00000076635 ENSRNOT00000068098 |
Snx12
|
sorting nexin 12 |
| chr10_-_56289882 | 3.08 |
ENSRNOT00000090762
ENSRNOT00000056903 |
Tnfsf13
|
tumor necrosis factor superfamily member 13 |
| chrX_+_70461718 | 3.06 |
ENSRNOT00000078233
ENSRNOT00000003789 |
Kif4a
|
kinesin family member 4A |
| chr7_+_63922879 | 3.06 |
ENSRNOT00000043581
|
RGD1565498
|
similar to Hypothetical protein LOC270802 |
| chr3_+_66673071 | 2.99 |
ENSRNOT00000034769
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr16_-_74264142 | 2.99 |
ENSRNOT00000026067
|
Dkk4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr15_-_34469350 | 2.96 |
ENSRNOT00000067536
|
Adcy4
|
adenylate cyclase 4 |
| chr5_+_16526058 | 2.96 |
ENSRNOT00000011130
|
Lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr5_-_136726358 | 2.90 |
ENSRNOT00000026739
|
Dph2
|
DPH2 homolog |
| chr15_-_32888095 | 2.88 |
ENSRNOT00000012233
|
Dad1
|
defender against cell death 1 |
| chr6_+_43363792 | 2.86 |
ENSRNOT00000091061
|
Cpsf3
|
cleavage and polyadenylation specific factor 3 |
| chr1_+_86972244 | 2.78 |
ENSRNOT00000036907
|
Rinl
|
Ras and Rab interactor-like |
| chr14_+_104475082 | 2.77 |
ENSRNOT00000084481
|
Rab1a
|
RAB1A, member RAS oncogene family |
| chr1_-_221015929 | 2.75 |
ENSRNOT00000028137
|
Sipa1
|
signal-induced proliferation-associated 1 |
| chr18_+_24397369 | 2.74 |
ENSRNOT00000022761
|
Sap130
|
Sin3A associated protein 130 |
| chr3_-_162579201 | 2.72 |
ENSRNOT00000068328
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr10_+_58342393 | 2.72 |
ENSRNOT00000010358
|
Wscd1
|
WSC domain containing 1 |
| chr1_+_221409271 | 2.69 |
ENSRNOT00000028462
|
Syvn1
|
synoviolin 1 |
| chr7_-_122581777 | 2.68 |
ENSRNOT00000025758
|
Slc25a17
|
solute carrier family 25 member 17 |
| chr10_-_13892997 | 2.61 |
ENSRNOT00000004192
|
Traf7
|
TNF receptor associated factor 7 |
| chr6_+_26836216 | 2.61 |
ENSRNOT00000077903
ENSRNOT00000011373 |
Ost4
|
oligosaccharyltransferase complex subunit 4, non-catalytic |
| chr10_+_85978691 | 2.60 |
ENSRNOT00000006359
|
Rpl19
|
ribosomal protein L19 |
| chr16_-_2367069 | 2.59 |
ENSRNOT00000079625
|
Pde12
|
phosphodiesterase 12 |
| chr5_+_154260062 | 2.56 |
ENSRNOT00000074998
|
Cnr2
|
cannabinoid receptor 2 |
| chr3_-_3890758 | 2.56 |
ENSRNOT00000064083
|
Sec16a
|
SEC16 homolog A, endoplasmic reticulum export factor |
| chr10_-_30118873 | 2.54 |
ENSRNOT00000006063
|
Ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr16_+_20825044 | 2.50 |
ENSRNOT00000027268
|
Upf1
|
UPF1, RNA helicase and ATPase |
| chr10_-_110585376 | 2.46 |
ENSRNOT00000054917
|
Rab40b
|
Rab40b, member RAS oncogene family |
| chr7_-_12085226 | 2.46 |
ENSRNOT00000088669
ENSRNOT00000079925 |
Onecut3
|
one cut domain, family member 3 |
| chr20_-_4818568 | 2.46 |
ENSRNOT00000001115
|
Ddx39b
|
DExD-box helicase 39B |
| chr10_+_59360765 | 2.44 |
ENSRNOT00000036278
|
Zzef1
|
zinc finger ZZ-type and EF-hand domain containing 1 |
| chr10_+_74002151 | 2.44 |
ENSRNOT00000005661
|
Ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr2_-_187786700 | 2.41 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr3_-_152222647 | 2.34 |
ENSRNOT00000026820
|
Nfs1
|
NFS1 cysteine desulfurase |
| chr1_-_222098297 | 2.27 |
ENSRNOT00000028678
|
Rps6ka4
|
ribosomal protein S6 kinase A4 |
| chr5_-_173653905 | 2.22 |
ENSRNOT00000038556
|
Plekhn1
|
pleckstrin homology domain containing N1 |
| chr1_-_82409639 | 2.22 |
ENSRNOT00000031326
|
Erich4
|
glutamate-rich 4 |
| chr2_-_189602143 | 2.15 |
ENSRNOT00000032062
|
Creb3l4
|
cAMP responsive element binding protein 3-like 4 |
| chr5_-_171710312 | 2.14 |
ENSRNOT00000076044
ENSRNOT00000071280 ENSRNOT00000072059 ENSRNOT00000077015 |
Prdm16
|
PR/SET domain 16 |
| chr1_+_12823363 | 2.14 |
ENSRNOT00000086790
|
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr6_+_104291340 | 2.13 |
ENSRNOT00000089313
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr1_+_142060955 | 2.10 |
ENSRNOT00000017862
|
Vps33b
|
VPS33B, late endosome and lysosome associated |
| chr2_-_78283473 | 2.03 |
ENSRNOT00000087246
|
AABR07008911.1
|
|
| chr13_+_35554964 | 2.03 |
ENSRNOT00000072632
|
Tmem185b
|
transmembrane protein 185B |
| chr7_-_60309878 | 2.00 |
ENSRNOT00000007610
|
RGD1306474
|
similar to RIKEN cDNA 9530003J23 |
| chr20_+_5646097 | 1.96 |
ENSRNOT00000090925
|
Itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr3_-_151688149 | 1.89 |
ENSRNOT00000072945
|
Nfs1
|
NFS1 cysteine desulfurase |
| chr7_+_12471824 | 1.89 |
ENSRNOT00000068197
|
Sbno2
|
strawberry notch homolog 2 |
| chr20_+_2094931 | 1.84 |
ENSRNOT00000001013
|
Ppp1r11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr18_+_40586315 | 1.83 |
ENSRNOT00000041237
|
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr1_-_7801438 | 1.82 |
ENSRNOT00000022273
|
AABR07000276.1
|
|
| chr13_+_56513286 | 1.80 |
ENSRNOT00000015596
|
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr9_+_9704346 | 1.77 |
ENSRNOT00000075403
|
Gpr108
|
G protein-coupled receptor 108 |
| chr10_+_55924938 | 1.68 |
ENSRNOT00000087003
ENSRNOT00000057079 |
Trappc1
|
trafficking protein particle complex 1 |
| chr3_+_122544788 | 1.68 |
ENSRNOT00000063828
|
Tgm3
|
transglutaminase 3 |
| chr8_+_49354115 | 1.66 |
ENSRNOT00000032837
|
Mpzl3
|
myelin protein zero-like 3 |
| chr10_+_89174684 | 1.58 |
ENSRNOT00000043754
|
Vps25
|
vacuolar protein sorting 25 |
| chr11_-_35749464 | 1.58 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chr8_-_55171718 | 1.58 |
ENSRNOT00000080736
|
LOC689959
|
hypothetical protein LOC689959 |
| chr5_+_120498883 | 1.53 |
ENSRNOT00000007859
|
Leprot
|
leptin receptor overlapping transcript |
| chr18_+_1971506 | 1.51 |
ENSRNOT00000092027
|
Mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr11_+_46184871 | 1.50 |
ENSRNOT00000048417
|
Tfg
|
Trk-fused gene |
| chr13_+_82231030 | 1.50 |
ENSRNOT00000003551
|
Scyl3
|
SCY1 like pseudokinase 3 |
| chr7_-_3246071 | 1.44 |
ENSRNOT00000044292
|
Ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr18_-_27159693 | 1.37 |
ENSRNOT00000027345
|
Reep5
|
receptor accessory protein 5 |
| chr17_-_63994169 | 1.37 |
ENSRNOT00000075651
|
Chrm3
|
cholinergic receptor, muscarinic 3 |
| chr17_-_90071408 | 1.34 |
ENSRNOT00000071978
|
Pdss1
|
decaprenyl diphosphate synthase subunit 1 |
| chr9_+_114022137 | 1.32 |
ENSRNOT00000007657
|
Map3k7
|
mitogen activated protein kinase kinase kinase 7 |
| chr19_-_37528011 | 1.31 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr18_-_40586260 | 1.30 |
ENSRNOT00000004942
|
Tmed7
|
transmembrane p24 trafficking protein 7 |
| chr10_+_43768708 | 1.28 |
ENSRNOT00000086218
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr17_-_10001901 | 1.25 |
ENSRNOT00000082836
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr7_-_140919234 | 1.23 |
ENSRNOT00000082470
|
Mcrs1
|
microspherule protein 1 |
| chr2_+_198006316 | 1.23 |
ENSRNOT00000081383
ENSRNOT00000056289 |
Aph1a
|
aph-1 homolog A, gamma secretase subunit |
| chr3_-_122703705 | 1.20 |
ENSRNOT00000079923
ENSRNOT00000049857 |
Snrpb
|
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr8_-_64154396 | 1.20 |
ENSRNOT00000031262
|
Bbs4
|
Bardet-Biedl syndrome 4 |
| chr10_+_46818525 | 1.16 |
ENSRNOT00000088120
|
Drg2
|
developmentally regulated GTP binding protein 2 |
| chr1_-_260517323 | 1.16 |
ENSRNOT00000018043
|
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr17_+_45078556 | 1.15 |
ENSRNOT00000088280
|
Zfp192
|
zinc finger protein 192 |
| chr1_-_65664767 | 1.14 |
ENSRNOT00000026528
ENSRNOT00000088367 |
Rps5
|
ribosomal protein S5 |
| chr17_+_81209867 | 1.11 |
ENSRNOT00000077368
ENSRNOT00000087129 |
Stam
|
signal transducing adaptor molecule |
| chr10_-_16792909 | 1.11 |
ENSRNOT00000004653
|
Atp6v0e1
|
ATPase H+ transporting V0 subunit e1 |
| chr1_+_221735517 | 1.09 |
ENSRNOT00000028628
ENSRNOT00000044866 |
Sf1
|
splicing factor 1 |
| chr5_-_139748489 | 1.04 |
ENSRNOT00000078741
|
Nfyc
|
nuclear transcription factor Y subunit gamma |
| chr5_-_140585408 | 1.04 |
ENSRNOT00000018711
|
Cap1
|
adenylate cyclase associated protein 1 |
| chr3_-_59688692 | 1.03 |
ENSRNOT00000078752
|
Sp3
|
Sp3 transcription factor |
| chr10_+_47018974 | 0.99 |
ENSRNOT00000079375
|
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr6_+_108936664 | 0.98 |
ENSRNOT00000007298
|
Dlst
|
dihydrolipoamide S-succinyltransferase |
| chr2_-_178389608 | 0.92 |
ENSRNOT00000013262
|
Etfdh
|
electron transfer flavoprotein dehydrogenase |
| chrX_+_10964067 | 0.91 |
ENSRNOT00000093181
ENSRNOT00000066480 |
Med14
|
mediator complex subunit 14 |
| chr18_+_25037625 | 0.90 |
ENSRNOT00000018422
|
Ercc3
|
ERCC excision repair 3, TFIIH core complex helicase subunit |
| chr12_+_47103314 | 0.90 |
ENSRNOT00000064910
|
Rnf10
|
ring finger protein 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Spdef
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.5 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 3.3 | 13.2 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 2.7 | 8.2 | GO:0002432 | granuloma formation(GO:0002432) |
| 2.7 | 10.9 | GO:0051692 | cellular oligosaccharide catabolic process(GO:0051692) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 2.5 | 9.8 | GO:0071226 | cellular response to molecule of fungal origin(GO:0071226) |
| 2.1 | 6.4 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 2.1 | 12.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 2.0 | 5.9 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 1.8 | 5.4 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.8 | 7.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 1.7 | 5.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 1.5 | 5.9 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 1.5 | 7.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 1.5 | 8.7 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 1.4 | 4.2 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 1.4 | 5.4 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 1.3 | 4.0 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 1.3 | 3.8 | GO:0060939 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 1.3 | 3.8 | GO:2000464 | positive regulation of astrocyte chemotaxis(GO:2000464) |
| 1.2 | 3.5 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 1.2 | 3.5 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 1.1 | 3.3 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 1.1 | 21.7 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 1.1 | 14.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 1.1 | 3.2 | GO:0035262 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) gonad morphogenesis(GO:0035262) nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 1.0 | 13.8 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 1.0 | 3.0 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.9 | 5.5 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.9 | 2.7 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.9 | 2.7 | GO:0043132 | NAD transport(GO:0043132) |
| 0.9 | 4.4 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.9 | 3.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.9 | 3.5 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 0.8 | 10.8 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.8 | 2.5 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.7 | 7.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.7 | 8.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.7 | 4.8 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.7 | 2.7 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.7 | 5.4 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.7 | 0.7 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.6 | 3.8 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.6 | 4.4 | GO:0051940 | regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) nuclear membrane organization(GO:0071763) |
| 0.6 | 1.9 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.6 | 3.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.6 | 3.1 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.6 | 2.9 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.6 | 2.3 | GO:0043988 | histone H3-S10 phosphorylation(GO:0043987) histone H3-S28 phosphorylation(GO:0043988) |
| 0.5 | 8.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.5 | 3.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.5 | 6.8 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.5 | 2.6 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.5 | 4.6 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.5 | 3.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.5 | 4.9 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.5 | 1.4 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.5 | 1.4 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.5 | 2.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.4 | 3.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.4 | 8.0 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.4 | 7.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.4 | 6.9 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.4 | 2.1 | GO:0061428 | embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.4 | 3.0 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.4 | 2.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.4 | 5.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) negative regulation of protein homooligomerization(GO:0032463) |
| 0.4 | 3.7 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.4 | 5.6 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.4 | 3.3 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.4 | 2.5 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.3 | 2.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 5.9 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.3 | 2.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.3 | 1.0 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.3 | 1.2 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.3 | 1.2 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.3 | 0.9 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.3 | 2.6 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.3 | 2.6 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.3 | 3.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.3 | 1.6 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.3 | 3.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.3 | 5.9 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.3 | 4.8 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.3 | 1.8 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 0.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 0.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 2.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 | 3.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 4.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 1.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 10.5 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.2 | 1.2 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 | 6.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.2 | 3.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 6.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 11.1 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.2 | 5.8 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.2 | 0.9 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.2 | 10.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 4.0 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.2 | 5.7 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 1.0 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 3.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.2 | 2.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 3.2 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.2 | 0.5 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.2 | 1.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.4 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.1 | 4.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.7 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.8 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 3.7 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.1 | 2.7 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 8.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 0.9 | GO:0033683 | UV protection(GO:0009650) nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 0.9 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 1.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.6 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 1.1 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.1 | 1.8 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.5 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.7 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 3.0 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 2.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.4 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.1 | 3.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 1.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 28.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 3.1 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 2.6 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 0.1 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 4.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 2.9 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 4.8 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 1.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 1.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 | 1.5 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 1.1 | GO:0034620 | endoplasmic reticulum unfolded protein response(GO:0030968) cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.7 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.8 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.0 | 1.0 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.3 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 1.7 | GO:0042303 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 0.4 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 3.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 2.7 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.4 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 1.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 1.1 | GO:0043200 | response to amino acid(GO:0043200) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 1.4 | 8.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.1 | 4.4 | GO:0044393 | microspike(GO:0044393) prominosome(GO:0071914) |
| 1.1 | 3.3 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 1.1 | 3.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 1.0 | 9.9 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 1.0 | 3.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.9 | 7.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.9 | 2.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.9 | 3.6 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.7 | 13.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.6 | 3.8 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.6 | 4.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.5 | 3.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.5 | 1.6 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.5 | 2.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.5 | 16.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.5 | 7.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 2.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.4 | 2.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.4 | 1.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.4 | 8.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 3.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.3 | 5.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 3.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.3 | 5.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 5.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 10.7 | GO:0000791 | euchromatin(GO:0000791) |
| 0.2 | 1.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.2 | 3.0 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 6.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 0.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 6.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 1.0 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 2.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 10.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 5.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 3.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 8.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 3.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 1.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 11.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 2.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 3.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 13.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.9 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 24.1 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 0.5 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 0.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.3 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 0.1 | 1.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 8.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 6.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 3.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 6.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 11.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 6.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 9.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 4.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 4.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 3.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.9 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 6.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.5 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 4.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 2.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.8 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 3.8 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 2.1 | 18.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 1.8 | 7.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) PTB domain binding(GO:0051425) |
| 1.7 | 6.6 | GO:0035877 | death effector domain binding(GO:0035877) |
| 1.5 | 7.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.4 | 5.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.3 | 3.8 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 1.2 | 3.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 1.1 | 7.8 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.1 | 8.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 1.1 | 6.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 1.1 | 8.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.1 | 3.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.9 | 3.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.9 | 5.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.8 | 5.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.8 | 8.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.8 | 3.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.8 | 5.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.7 | 15.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.7 | 6.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.7 | 13.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.6 | 2.6 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.6 | 2.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.6 | 12.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.6 | 10.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.5 | 9.8 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.5 | 12.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.5 | 2.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.5 | 7.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.5 | 4.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.5 | 1.4 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.5 | 4.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.4 | 5.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.4 | 3.6 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.4 | 1.2 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.4 | 3.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.4 | 4.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.4 | 11.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.4 | 10.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 3.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.3 | 5.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 7.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.3 | 3.0 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.3 | 5.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 1.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.3 | 3.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 2.5 | GO:0030621 | U6 snRNA binding(GO:0017070) U4 snRNA binding(GO:0030621) |
| 0.3 | 1.5 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.3 | 2.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 0.9 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.3 | 4.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 14.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 0.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 8.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 10.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 5.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 9.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 2.9 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 1.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 3.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 10.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 1.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 0.9 | GO:0032564 | dATP binding(GO:0032564) |
| 0.2 | 1.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) telomerase inhibitor activity(GO:0010521) |
| 0.2 | 1.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 7.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 3.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.2 | 1.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 2.0 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 1.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 2.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.8 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.1 | 4.4 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 3.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 2.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 3.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 8.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 4.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 3.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 23.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 7.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 2.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 4.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 0.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 3.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 5.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 2.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 4.2 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 4.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 2.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 3.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 6.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 5.5 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 4.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 13.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 12.1 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 5.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 12.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.3 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 2.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.7 | 6.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.7 | 13.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.7 | 40.7 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.6 | 6.8 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.5 | 8.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 4.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 3.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 1.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 11.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 11.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.2 | 3.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 4.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 14.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 4.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 6.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 3.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 2.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 5.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 3.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 18.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 1.0 | 3.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.6 | 12.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.6 | 6.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.5 | 11.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.5 | 8.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.4 | 6.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.4 | 5.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 4.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 5.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.3 | 3.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.3 | 10.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.3 | 8.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.2 | 5.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 3.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 3.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 11.6 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.2 | 2.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 27.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 4.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 5.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 1.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 2.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 1.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 7.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 4.1 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.1 | 2.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 3.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 3.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.6 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.1 | 1.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 6.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 4.0 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 7.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 3.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.8 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.4 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |