Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
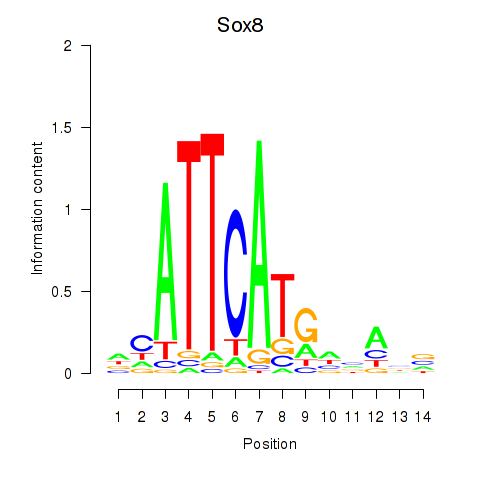
Results for Sox8
Z-value: 0.35
Transcription factors associated with Sox8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox8
|
ENSRNOG00000018841 | SRY box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox8 | rn6_v1_chr10_-_14937336_14937336 | 0.57 | 4.9e-29 | Click! |
Activity profile of Sox8 motif
Sorted Z-values of Sox8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_39305128 | 12.03 |
ENSRNOT00000008285
|
Fez1
|
fasciculation and elongation protein zeta 1 |
| chr9_-_75528644 | 11.92 |
ENSRNOT00000019283
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chr16_-_18937562 | 9.35 |
ENSRNOT00000080387
|
Nwd1
|
NACHT and WD repeat domain containing 1 |
| chr1_-_198233588 | 9.03 |
ENSRNOT00000088473
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr17_+_53231343 | 7.93 |
ENSRNOT00000021703
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr18_+_15490717 | 7.85 |
ENSRNOT00000091557
|
B4galt6
|
beta-1,4-galactosyltransferase 6 |
| chr17_-_52477575 | 7.84 |
ENSRNOT00000081290
|
Gli3
|
GLI family zinc finger 3 |
| chr1_-_198233215 | 7.81 |
ENSRNOT00000087928
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr1_+_224882439 | 7.57 |
ENSRNOT00000024785
|
Chrm1
|
cholinergic receptor, muscarinic 1 |
| chr4_-_150485781 | 6.62 |
ENSRNOT00000008763
|
Zfp248
|
zinc finger protein 248 |
| chr1_+_99486253 | 4.29 |
ENSRNOT00000074820
|
LOC102546648
|
uncharacterized LOC102546648 |
| chr18_+_17403407 | 4.09 |
ENSRNOT00000045150
|
RGD1562608
|
similar to KIAA1328 protein |
| chr15_+_41937880 | 3.95 |
ENSRNOT00000032514
|
Kcnrg
|
potassium channel regulator |
| chr5_-_819326 | 3.89 |
ENSRNOT00000023899
|
Pi15
|
peptidase inhibitor 15 |
| chr20_+_1787174 | 3.81 |
ENSRNOT00000072824
ENSRNOT00000041883 |
Olr1738
|
olfactory receptor 1738 |
| chr6_-_132972511 | 3.61 |
ENSRNOT00000082216
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr1_+_99505677 | 2.88 |
ENSRNOT00000024645
|
Zfp719
|
zinc finger protein 719 |
| chr7_+_44009069 | 2.76 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr12_+_22259713 | 2.75 |
ENSRNOT00000001913
|
Pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr7_-_140437467 | 2.75 |
ENSRNOT00000087181
|
Fkbp11
|
FK506 binding protein 11 |
| chr20_+_27954433 | 2.24 |
ENSRNOT00000064288
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr7_-_15821927 | 2.06 |
ENSRNOT00000050658
|
LOC691422
|
similar to zinc finger protein 101 |
| chr2_+_22909569 | 2.04 |
ENSRNOT00000073871
|
Homer1
|
homer scaffolding protein 1 |
| chr2_-_117454769 | 1.80 |
ENSRNOT00000068381
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr8_+_18992899 | 1.55 |
ENSRNOT00000061326
|
Olr1129
|
olfactory receptor 1129 |
| chr1_+_170109291 | 1.37 |
ENSRNOT00000041317
|
Olr198
|
olfactory receptor 198 |
| chr1_+_88451958 | 1.34 |
ENSRNOT00000074301
|
LOC102549115
|
zinc finger protein 260-like |
| chr1_-_80221417 | 1.32 |
ENSRNOT00000072149
|
Fosb
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr1_-_80221710 | 1.19 |
ENSRNOT00000091687
|
Fosb
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr13_+_52976507 | 1.08 |
ENSRNOT00000090599
ENSRNOT00000011324 |
Kif21b
|
kinesin family member 21B |
| chr2_-_250600517 | 0.96 |
ENSRNOT00000016872
|
Hs2st1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr18_+_76559811 | 0.73 |
ENSRNOT00000084621
|
Pard6g
|
par-6 family cell polarity regulator gamma |
| chr16_-_20641908 | 0.61 |
ENSRNOT00000026846
|
Ell
|
elongation factor for RNA polymerase II |
| chr1_+_170147300 | 0.54 |
ENSRNOT00000048075
|
Olr200
|
olfactory receptor 200 |
| chr13_-_98529040 | 0.53 |
ENSRNOT00000091715
|
Psen2
|
presenilin 2 |
| chr9_+_65110330 | 0.37 |
ENSRNOT00000033068
ENSRNOT00000092868 |
Aox4
|
aldehyde oxidase 4 |
| chr1_+_230217215 | 0.34 |
ENSRNOT00000072772
|
LOC687088
|
similar to olfactory receptor 1467 |
| chr1_-_224986291 | 0.28 |
ENSRNOT00000026284
|
Taf6l
|
TATA-box binding protein associated factor 6 like |
| chr8_-_60752047 | 0.27 |
ENSRNOT00000048115
|
AABR07070238.2
|
|
| chr1_+_169616178 | 0.26 |
ENSRNOT00000023170
|
Olr157
|
olfactory receptor 157 |
| chr11_+_43699020 | 0.06 |
ENSRNOT00000070849
|
Olr1559
|
olfactory receptor 1559 |
| chr11_+_28780446 | 0.05 |
ENSRNOT00000072546
|
Krtap15-1
|
keratin associated protein 15-1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 7.8 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 1.5 | 7.6 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.2 | 9.9 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 1.2 | 16.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.0 | 12.0 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.5 | 7.9 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.3 | 1.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 4.0 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.3 | 2.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 | 1.8 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 2.0 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.6 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.4 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 2.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 2.5 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 2.8 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 7.9 | GO:0016567 | protein ubiquitination(GO:0016567) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 16.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 7.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 7.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 0.6 | GO:0035363 | histone locus body(GO:0035363) |
| 0.1 | 7.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 11.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.0 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 16.7 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 16.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 2.0 | 7.9 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 1.5 | 7.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.3 | 11.9 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.4 | 2.0 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.3 | 12.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 2.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 7.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.4 | GO:0004031 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.1 | 1.0 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 2.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 7.9 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 2.8 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 1.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 16.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 7.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.4 | 11.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.4 | 16.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 7.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.5 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |