Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
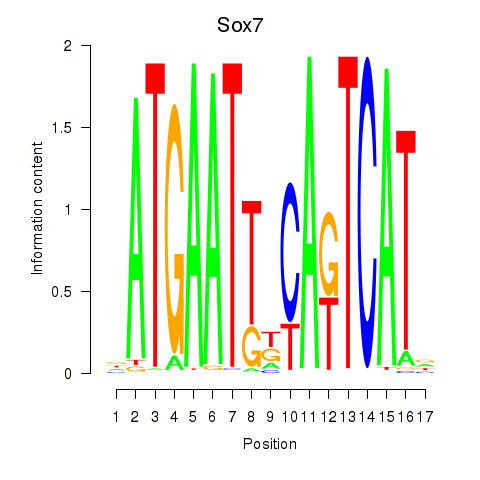
Results for Sox7
Z-value: 0.49
Transcription factors associated with Sox7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox7
|
ENSRNOG00000012049 | SRY box 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox7 | rn6_v1_chr15_+_47293699_47293699 | -0.09 | 9.1e-02 | Click! |
Activity profile of Sox7 motif
Sorted Z-values of Sox7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_75481115 | 15.43 |
ENSRNOT00000035128
|
Defa7
|
defensin alpha 7 |
| chr16_-_75340360 | 13.61 |
ENSRNOT00000018501
|
Defa5
|
defensin alpha 5 |
| chr18_+_25749098 | 11.32 |
ENSRNOT00000027771
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr15_+_30270740 | 9.42 |
ENSRNOT00000070991
|
AABR07017707.1
|
|
| chr9_-_84894599 | 9.24 |
ENSRNOT00000018423
|
Hdac1l
|
histone deacetylase 1-like |
| chr15_+_31948035 | 8.65 |
ENSRNOT00000071627
|
AABR07017868.1
|
|
| chr18_+_63599425 | 7.45 |
ENSRNOT00000023145
|
Cep192
|
centrosomal protein 192 |
| chr10_-_49192693 | 7.42 |
ENSRNOT00000004294
|
Zfp286a
|
zinc finger protein 286A |
| chr15_-_29532988 | 7.07 |
ENSRNOT00000074782
|
AABR07017639.1
|
|
| chr1_+_32221636 | 6.47 |
ENSRNOT00000022346
ENSRNOT00000089941 |
Slc6a18
|
solute carrier family 6 member 18 |
| chr1_+_201672528 | 6.46 |
ENSRNOT00000093490
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr1_+_199596024 | 5.28 |
ENSRNOT00000085336
|
Itgad
|
integrin subunit alpha D |
| chr3_-_121882726 | 5.24 |
ENSRNOT00000006308
|
Il1b
|
interleukin 1 beta |
| chr14_+_91557601 | 5.12 |
ENSRNOT00000038733
|
RGD1309870
|
hypothetical LOC289778 |
| chr2_+_60920257 | 4.65 |
ENSRNOT00000025170
|
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr5_-_58950373 | 4.63 |
ENSRNOT00000060442
|
Cd72
|
Cd72 molecule |
| chr3_+_2504694 | 4.34 |
ENSRNOT00000061987
|
Tmem210
|
transmembrane protein 210 |
| chr15_+_31579478 | 4.17 |
ENSRNOT00000071642
|
AABR07017830.1
|
|
| chr17_-_14373983 | 4.17 |
ENSRNOT00000071112
|
AABR07027098.1
|
|
| chr20_-_32139789 | 4.14 |
ENSRNOT00000078140
|
Srgn
|
serglycin |
| chr5_+_131920155 | 4.12 |
ENSRNOT00000064863
|
Skint1
|
selection and upkeep of intraepithelial T cells 1 |
| chr6_-_114476723 | 4.02 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr17_-_10622226 | 3.90 |
ENSRNOT00000044559
|
Simc1
|
SUMO-interacting motifs containing 1 |
| chr17_-_14949064 | 3.81 |
ENSRNOT00000079489
ENSRNOT00000081454 |
AABR07027128.1
|
|
| chr9_+_27343853 | 3.71 |
ENSRNOT00000072794
|
Tmem14a
|
transmembrane protein 14A |
| chrX_+_144994139 | 3.65 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chrX_-_106066227 | 3.62 |
ENSRNOT00000033752
|
Nxf7
|
nuclear RNA export factor 7 |
| chr3_-_8766433 | 3.52 |
ENSRNOT00000021865
|
Kyat1
|
kynurenine aminotransferase 1 |
| chr20_+_13498926 | 3.43 |
ENSRNOT00000070992
ENSRNOT00000045375 |
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr1_+_97777685 | 3.23 |
ENSRNOT00000030895
|
LOC690284
|
similar to F49E2.5d |
| chr5_+_4393472 | 3.03 |
ENSRNOT00000082717
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr16_+_52024690 | 2.95 |
ENSRNOT00000048104
|
AABR07025825.1
|
|
| chr10_+_17261541 | 2.94 |
ENSRNOT00000005478
|
Sh3pxd2b
|
SH3 and PX domains 2B |
| chr7_+_40316639 | 2.90 |
ENSRNOT00000080150
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr19_+_22569999 | 2.78 |
ENSRNOT00000022573
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr3_+_102456938 | 2.77 |
ENSRNOT00000051827
|
Olr748
|
olfactory receptor 748 |
| chrX_-_82418425 | 2.72 |
ENSRNOT00000022525
|
Ankra2
|
ankyrin repeat family A member 2 |
| chrX_-_153878806 | 2.67 |
ENSRNOT00000087990
|
Aff2
|
AF4/FMR2 family, member 2 |
| chr19_+_37852659 | 2.62 |
ENSRNOT00000030967
|
Nrn1l
|
neuritin 1-like |
| chr19_+_37852833 | 2.61 |
ENSRNOT00000006345
|
Nrn1l
|
neuritin 1-like |
| chr2_-_78427751 | 2.60 |
ENSRNOT00000077768
|
AC113901.1
|
|
| chr11_-_2813912 | 2.50 |
ENSRNOT00000070883
|
RGD1561853
|
similar to SET domain-containing protein |
| chr15_+_30440677 | 2.43 |
ENSRNOT00000085402
|
AABR07017737.1
|
|
| chr6_+_18880737 | 2.23 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr6_-_135251073 | 2.08 |
ENSRNOT00000088986
|
Mok
|
MOK protein kinase |
| chr1_+_101412736 | 2.00 |
ENSRNOT00000067468
|
Lhb
|
luteinizing hormone beta polypeptide |
| chrX_+_114930457 | 1.83 |
ENSRNOT00000089141
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr6_+_26566494 | 1.83 |
ENSRNOT00000079292
|
Gtf3c2
|
general transcription factor IIIC subunit 2 |
| chr20_+_1837976 | 1.75 |
ENSRNOT00000084564
|
Olr1742
|
olfactory receptor 1742 |
| chr4_+_102643934 | 1.67 |
ENSRNOT00000058389
|
LOC100911032
|
uncharacterized LOC100911032 |
| chrX_-_107442878 | 1.65 |
ENSRNOT00000052302
|
Glra4
|
glycine receptor, alpha 4 |
| chr10_+_39850818 | 1.62 |
ENSRNOT00000012671
|
Fnip1
|
folliculin interacting protein 1 |
| chr3_-_6054483 | 1.60 |
ENSRNOT00000084491
|
Brd3
|
bromodomain containing 3 |
| chr18_+_30487264 | 1.57 |
ENSRNOT00000040125
|
Pcdhb10
|
protocadherin beta 10 |
| chr4_-_103050006 | 1.42 |
ENSRNOT00000092130
|
LOC100911032
|
uncharacterized LOC100911032 |
| chr10_+_45798797 | 1.31 |
ENSRNOT00000039012
|
Jmjd4
|
jumonji domain containing 4 |
| chr1_-_73399377 | 1.30 |
ENSRNOT00000038898
|
Lilrb4
|
leukocyte immunoglobulin like receptor B4 |
| chrX_+_57870445 | 1.28 |
ENSRNOT00000065038
|
Ppp4r3c
|
protein phosphatase 4 regulatory subunit 3C |
| chr20_-_7482747 | 1.19 |
ENSRNOT00000038195
|
Taf11
|
TATA-box binding protein associated factor 11 |
| chr1_+_167758636 | 1.18 |
ENSRNOT00000024957
|
Olr46
|
olfactory receptor 46 |
| chr1_-_104024682 | 1.16 |
ENSRNOT00000056081
|
Mrgprx1
|
MAS-related GPR, member X1 |
| chr3_+_138398011 | 1.15 |
ENSRNOT00000038865
|
Mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr7_-_104486710 | 1.15 |
ENSRNOT00000084764
|
Gsdmc
|
gasdermin C |
| chr3_+_102471249 | 1.13 |
ENSRNOT00000051083
|
Olr749
|
olfactory receptor 749 |
| chr3_+_20007192 | 1.13 |
ENSRNOT00000075229
|
AABR07051716.1
|
|
| chr4_-_87982729 | 1.08 |
ENSRNOT00000089498
|
Vom1r79
|
vomeronasal 1 receptor 79 |
| chr8_+_113939886 | 1.07 |
ENSRNOT00000079562
|
Aste1
|
asteroid homolog 1 (Drosophila) |
| chr1_+_224927764 | 1.01 |
ENSRNOT00000050814
|
AC099294.1
|
|
| chr17_-_69460321 | 0.99 |
ENSRNOT00000058367
|
Akr1c1
|
aldo-keto reductase family 1, member C1 |
| chr11_+_43161181 | 0.89 |
ENSRNOT00000071470
|
Olr1530
|
olfactory receptor 1530 |
| chr4_+_31333970 | 0.88 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr9_-_17880706 | 0.87 |
ENSRNOT00000031549
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr11_+_72044096 | 0.86 |
ENSRNOT00000034757
|
Senp5
|
SUMO1/sentrin specific peptidase 5 |
| chr9_-_44560837 | 0.78 |
ENSRNOT00000090870
|
Mitd1
|
microtubule interacting and trafficking domain containing 1 |
| chr2_+_194557986 | 0.78 |
ENSRNOT00000070843
|
LOC100911679
|
TD and POZ domain-containing protein 2-like |
| chr1_+_101055622 | 0.77 |
ENSRNOT00000079189
|
Nosip
|
nitric oxide synthase interacting protein |
| chr10_-_13814304 | 0.75 |
ENSRNOT00000012203
|
Dnase1l2
|
deoxyribonuclease 1 like 2 |
| chr4_+_149908375 | 0.71 |
ENSRNOT00000019504
|
LOC100909657
|
uncharacterized LOC100909657 |
| chr9_-_44483655 | 0.60 |
ENSRNOT00000024959
|
Mitd1
|
microtubule interacting and trafficking domain containing 1 |
| chr1_-_60407295 | 0.54 |
ENSRNOT00000078350
|
Vom1r12
|
vomeronasal 1 receptor 12 |
| chr4_+_155531906 | 0.54 |
ENSRNOT00000060937
|
Nanog
|
Nanog homeobox |
| chr11_+_43329700 | 0.53 |
ENSRNOT00000060892
|
Olr1540
|
olfactory receptor 1540 |
| chr3_-_74545241 | 0.53 |
ENSRNOT00000090836
|
Olr531
|
olfactory receptor 531 |
| chr8_-_119523964 | 0.45 |
ENSRNOT00000081718
|
Mlh1
|
mutL homolog 1 |
| chr1_-_60281386 | 0.41 |
ENSRNOT00000092184
|
Vom1r10
|
vomeronasal 1 receptor 10 |
| chr2_-_34313094 | 0.41 |
ENSRNOT00000016863
|
Ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr1_-_168015148 | 0.40 |
ENSRNOT00000020691
|
Olr50
|
olfactory receptor 50 |
| chr3_+_148150698 | 0.38 |
ENSRNOT00000087075
ENSRNOT00000010251 |
Hm13
|
histocompatibility minor 13 |
| chr16_+_55152748 | 0.36 |
ENSRNOT00000000121
|
Fgf20
|
fibroblast growth factor 20 |
| chr7_-_40316532 | 0.30 |
ENSRNOT00000083347
|
RGD1307947
|
similar to RIKEN cDNA C430008C19 |
| chrX_+_158569056 | 0.26 |
ENSRNOT00000074170
|
LOC102550080
|
placenta-specific protein 1-like |
| chr4_-_167089055 | 0.18 |
ENSRNOT00000050409
|
Tas2r113
|
taste receptor, type 2, member 113 |
| chr4_-_87873247 | 0.06 |
ENSRNOT00000044351
|
Vom1r77
|
vomeronasal 1 receptor 77 |
| chr1_+_229066045 | 0.00 |
ENSRNOT00000016454
|
Glyat
|
glycine-N-acyltransferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 1.4 | 4.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.9 | 2.8 | GO:0042197 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.9 | 11.3 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.7 | 3.5 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.7 | 4.7 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.6 | 13.6 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.6 | 5.3 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.5 | 2.7 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.5 | 3.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.5 | 1.0 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.5 | 2.9 | GO:0002051 | osteoblast fate commitment(GO:0002051) positive regulation of adipose tissue development(GO:1904179) |
| 0.4 | 4.0 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) hormone catabolic process(GO:0042447) |
| 0.4 | 9.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.3 | 6.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 4.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.3 | 1.6 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.3 | 3.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.2 | 2.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 0.9 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 0.4 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) |
| 0.1 | 1.6 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 6.5 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 1.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.4 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 3.4 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 1.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 6.4 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.1 | 0.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 1.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 1.8 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 1.3 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 2.0 | GO:0030823 | regulation of cGMP metabolic process(GO:0030823) |
| 0.1 | 0.8 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 0.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 7.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.3 | 1.8 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 5.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.4 | GO:0005712 | chiasma(GO:0005712) recombination nodule(GO:0005713) |
| 0.1 | 11.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 6.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 2.1 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 4.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.4 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 5.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 1.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 3.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 3.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.9 | 11.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.9 | 6.5 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.7 | 9.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.6 | 2.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 3.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 3.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.3 | 2.7 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.3 | 1.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.3 | 2.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 5.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 7.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 6.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.2 | 1.6 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 1.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 0.9 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 1.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 2.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 2.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 1.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 4.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 5.3 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 3.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 4.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.6 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 14.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.3 | 5.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 5.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 4.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 6.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 4.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 13.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.3 | 11.3 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.3 | 6.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 4.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 2.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 7.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 4.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 5.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 4.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 1.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |