Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
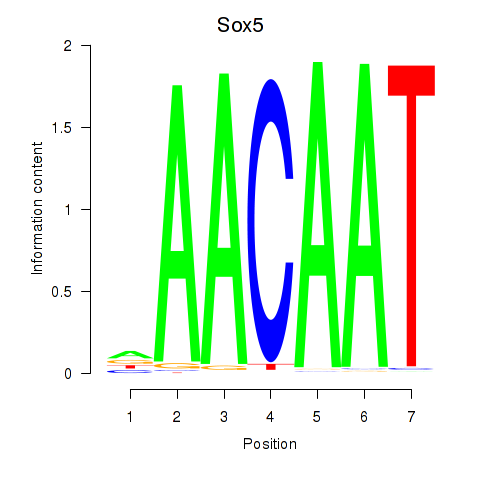
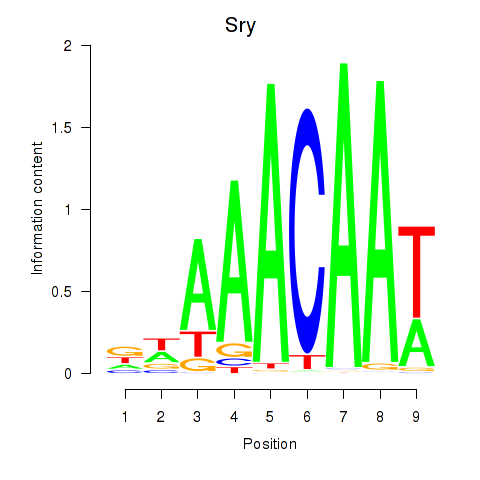
Results for Sox5_Sry
Z-value: 0.73
Transcription factors associated with Sox5_Sry
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox5
|
ENSRNOG00000027869 | SRY box 5 |
|
Sry
|
ENSRNOG00000062090 | sex determining region Y |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sry | rn6_v1_chrY_+_327166_327166 | 0.12 | 2.9e-02 | Click! |
| Sox5 | rn6_v1_chr4_-_178441547_178441547 | 0.07 | 2.4e-01 | Click! |
Activity profile of Sox5_Sry motif
Sorted Z-values of Sox5_Sry motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_72219246 | 29.67 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr7_+_38858062 | 26.84 |
ENSRNOT00000006234
|
Kera
|
keratocan |
| chr18_-_26211445 | 17.93 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr4_-_14490446 | 14.42 |
ENSRNOT00000009132
|
Sema3c
|
semaphorin 3C |
| chr4_+_44774741 | 13.74 |
ENSRNOT00000086902
|
Met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr9_-_53315915 | 12.88 |
ENSRNOT00000038093
|
Mstn
|
myostatin |
| chr18_+_79406381 | 10.47 |
ENSRNOT00000022303
ENSRNOT00000058295 ENSRNOT00000058296 ENSRNOT00000022280 |
Mbp
|
myelin basic protein |
| chr11_+_58624198 | 10.35 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chrX_+_62727755 | 9.93 |
ENSRNOT00000077055
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr11_+_61083757 | 9.74 |
ENSRNOT00000002790
|
Boc
|
BOC cell adhesion associated, oncogene regulated |
| chr9_+_37727942 | 9.39 |
ENSRNOT00000016511
ENSRNOT00000074276 |
LOC100912306
|
myotilin-like |
| chr18_+_35574002 | 9.37 |
ENSRNOT00000093442
ENSRNOT00000070817 ENSRNOT00000093356 |
Myot
|
myotilin |
| chr16_-_6245644 | 9.23 |
ENSRNOT00000040759
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr2_-_172361779 | 8.84 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr5_+_64326733 | 7.82 |
ENSRNOT00000065775
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr16_+_3293599 | 7.79 |
ENSRNOT00000081999
ENSRNOT00000047118 ENSRNOT00000020427 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr13_-_68360664 | 7.64 |
ENSRNOT00000030971
|
Hmcn1
|
hemicentin 1 |
| chr1_-_93949187 | 7.51 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr7_+_142776252 | 6.98 |
ENSRNOT00000008673
|
Acvrl1
|
activin A receptor like type 1 |
| chr7_+_142776580 | 6.52 |
ENSRNOT00000081047
|
Acvrl1
|
activin A receptor like type 1 |
| chr6_-_125723944 | 6.31 |
ENSRNOT00000007004
|
Fbln5
|
fibulin 5 |
| chr14_+_114126943 | 6.19 |
ENSRNOT00000041638
ENSRNOT00000006443 ENSRNOT00000006957 |
Rtn4
|
reticulon 4 |
| chr4_+_155321553 | 6.16 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr4_-_119131202 | 6.05 |
ENSRNOT00000011675
|
Antxr1
|
anthrax toxin receptor 1 |
| chr2_-_113616766 | 5.78 |
ENSRNOT00000016858
ENSRNOT00000074723 |
Tmem212
|
transmembrane protein 212 |
| chr1_-_126211439 | 5.73 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr4_-_39102807 | 5.67 |
ENSRNOT00000052063
|
Thsd7a
|
thrombospondin type 1 domain containing 7A |
| chr7_-_138483612 | 5.54 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr1_+_256955652 | 5.52 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
| chr2_+_242634399 | 5.24 |
ENSRNOT00000035700
|
Emcn
|
endomucin |
| chr3_-_123718432 | 5.23 |
ENSRNOT00000028861
|
Spef1
|
sperm flagellar 1 |
| chr11_-_35749464 | 5.17 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chr3_-_51643140 | 5.02 |
ENSRNOT00000006646
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr5_+_104362971 | 5.00 |
ENSRNOT00000058520
|
Adamtsl1
|
ADAMTS-like 1 |
| chr13_-_112099336 | 4.73 |
ENSRNOT00000009158
ENSRNOT00000044161 |
Camk1g
|
calcium/calmodulin-dependent protein kinase IG |
| chr9_-_55256340 | 4.45 |
ENSRNOT00000028907
|
Sdpr
|
serum deprivation response |
| chr15_+_83703791 | 4.44 |
ENSRNOT00000090637
|
Klf5
|
Kruppel-like factor 5 |
| chr18_-_5314511 | 4.43 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr2_-_231521052 | 4.41 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr6_-_125723732 | 4.11 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr6_+_27768943 | 4.04 |
ENSRNOT00000015820
|
Kif3c
|
kinesin family member 3C |
| chr16_+_46731403 | 4.02 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr1_+_234252757 | 3.89 |
ENSRNOT00000091814
|
Rorb
|
RAR-related orphan receptor B |
| chr18_-_38088457 | 3.87 |
ENSRNOT00000077814
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr4_-_108008484 | 3.79 |
ENSRNOT00000007971
|
Ctnna2
|
catenin alpha 2 |
| chr6_-_67084234 | 3.74 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr11_+_9642365 | 3.72 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chrX_+_9436707 | 3.59 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr6_+_110624856 | 3.55 |
ENSRNOT00000014017
|
Vash1
|
vasohibin 1 |
| chr8_-_84522588 | 3.43 |
ENSRNOT00000076213
|
Mlip
|
muscular LMNA-interacting protein |
| chr8_+_23113048 | 3.43 |
ENSRNOT00000029577
|
Cnn1
|
calponin 1 |
| chr2_-_35104963 | 3.35 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr2_-_63166509 | 3.27 |
ENSRNOT00000018246
|
Cdh6
|
cadherin 6 |
| chrX_+_40363646 | 3.27 |
ENSRNOT00000010135
|
Sms
|
spermine synthase |
| chr6_-_106971250 | 3.18 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr3_-_168018410 | 3.00 |
ENSRNOT00000087579
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr15_+_33600102 | 2.99 |
ENSRNOT00000022664
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr4_-_55398941 | 2.92 |
ENSRNOT00000075324
|
Grm8
|
glutamate metabotropic receptor 8 |
| chrX_-_115175299 | 2.88 |
ENSRNOT00000074322
|
Dcx
|
doublecortin |
| chr4_-_148711830 | 2.87 |
ENSRNOT00000049091
|
Olr832
|
olfactory receptor 832 |
| chr13_-_111581018 | 2.87 |
ENSRNOT00000083072
ENSRNOT00000077981 |
Sertad4
|
SERTA domain containing 4 |
| chr1_-_90520344 | 2.86 |
ENSRNOT00000078598
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr7_+_116632506 | 2.85 |
ENSRNOT00000009811
|
Gpihbp1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr2_+_203768117 | 2.85 |
ENSRNOT00000080176
|
Igsf3
|
immunoglobulin superfamily, member 3 |
| chr18_+_30381322 | 2.81 |
ENSRNOT00000027216
|
Pcdhb3
|
protocadherin beta 3 |
| chr18_+_3887419 | 2.76 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr1_+_241594565 | 2.71 |
ENSRNOT00000020123
|
Apba1
|
amyloid beta precursor protein binding family A member 1 |
| chr10_+_23661013 | 2.70 |
ENSRNOT00000076664
|
Ebf1
|
early B-cell factor 1 |
| chr10_-_46720907 | 2.68 |
ENSRNOT00000083093
ENSRNOT00000067866 |
Tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr15_+_33600337 | 2.54 |
ENSRNOT00000075965
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr7_+_59326518 | 2.49 |
ENSRNOT00000085231
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr9_+_67234303 | 2.49 |
ENSRNOT00000050179
|
Abi2
|
abl-interactor 2 |
| chr6_-_136436620 | 2.42 |
ENSRNOT00000067118
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chr15_-_93307420 | 2.41 |
ENSRNOT00000012195
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr16_-_71319449 | 2.36 |
ENSRNOT00000029284
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr8_-_90664554 | 2.26 |
ENSRNOT00000039917
ENSRNOT00000074896 ENSRNOT00000072225 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr2_-_187401786 | 2.24 |
ENSRNOT00000025624
|
Hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr5_-_12563429 | 2.20 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr3_+_137618898 | 2.18 |
ENSRNOT00000007249
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr5_+_113725717 | 2.17 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr6_-_136436818 | 2.13 |
ENSRNOT00000082600
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chr3_+_116899878 | 2.13 |
ENSRNOT00000090802
ENSRNOT00000066101 |
Sema6d
|
semaphorin 6D |
| chr3_+_35014538 | 2.11 |
ENSRNOT00000006341
|
Kif5c
|
kinesin family member 5C |
| chr4_+_158088505 | 2.10 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr11_-_17684903 | 2.09 |
ENSRNOT00000051213
|
Tmprss15
|
transmembrane protease, serine 15 |
| chr10_-_37455022 | 2.08 |
ENSRNOT00000016742
|
LOC103694902
|
ubiquitin-conjugating enzyme E2 B |
| chr16_-_71319052 | 2.06 |
ENSRNOT00000050980
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr13_-_95250235 | 2.04 |
ENSRNOT00000085648
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr16_-_47535358 | 2.03 |
ENSRNOT00000040731
|
Cldn22
|
claudin 22 |
| chr10_+_14248399 | 2.00 |
ENSRNOT00000077689
|
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr10_-_34439470 | 1.96 |
ENSRNOT00000072081
|
Btnl9
|
butyrophilin-like 9 |
| chr8_+_14811627 | 1.93 |
ENSRNOT00000043392
|
LOC689840
|
LRRGT00142 |
| chrX_+_6273733 | 1.92 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr18_+_65155685 | 1.89 |
ENSRNOT00000081797
|
Tcf4
|
transcription factor 4 |
| chr14_+_3882398 | 1.83 |
ENSRNOT00000033794
|
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog |
| chr10_+_43768708 | 1.78 |
ENSRNOT00000086218
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr4_-_115516296 | 1.74 |
ENSRNOT00000019399
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chr3_-_71845232 | 1.73 |
ENSRNOT00000078645
|
Calcrl
|
calcitonin receptor like receptor |
| chr16_+_23668595 | 1.72 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_-_206282575 | 1.69 |
ENSRNOT00000024974
|
Adam12
|
ADAM metallopeptidase domain 12 |
| chr14_+_17492081 | 1.66 |
ENSRNOT00000003346
|
G3bp2
|
G3BP stress granule assembly factor 2 |
| chr1_+_144070754 | 1.62 |
ENSRNOT00000079989
|
Sh3gl3
|
SH3 domain-containing GRB2-like 3 |
| chr8_-_71337746 | 1.60 |
ENSRNOT00000021554
|
Zfp609
|
zinc finger protein 609 |
| chr6_-_42616548 | 1.58 |
ENSRNOT00000081433
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr6_+_91595823 | 1.54 |
ENSRNOT00000006223
|
Klhdc2
|
kelch domain containing 2 |
| chr6_+_73358112 | 1.48 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr10_+_67862054 | 1.47 |
ENSRNOT00000031746
|
Cdk5r1
|
cyclin-dependent kinase 5 regulatory subunit 1 |
| chr12_-_10335499 | 1.44 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chr9_-_5329305 | 1.42 |
ENSRNOT00000078055
|
Slc5a7
|
solute carrier family 5 member 7 |
| chr20_+_27592379 | 1.41 |
ENSRNOT00000047000
|
Trappc3l
|
trafficking protein particle complex 3-like |
| chr1_-_261090437 | 1.39 |
ENSRNOT00000072055
|
Frat2
|
FRAT2, WNT signaling pathway regulator |
| chr20_+_38935820 | 1.38 |
ENSRNOT00000001059
|
Hsf2
|
heat shock transcription factor 2 |
| chr4_+_8166082 | 1.38 |
ENSRNOT00000076429
|
Srpk2
|
SRSF protein kinase 2 |
| chr10_-_87261717 | 1.38 |
ENSRNOT00000015740
|
Krt27
|
keratin 27 |
| chr17_+_18151982 | 1.38 |
ENSRNOT00000066447
|
Kif13a
|
kinesin family member 13A |
| chr3_+_159569363 | 1.37 |
ENSRNOT00000064159
|
Tox2
|
TOX high mobility group box family member 2 |
| chr8_+_119030875 | 1.36 |
ENSRNOT00000028458
|
Myl3
|
myosin light chain 3 |
| chr7_+_140315368 | 1.29 |
ENSRNOT00000081206
|
Cacnb3
|
calcium voltage-gated channel auxiliary subunit beta 3 |
| chr10_+_66732390 | 1.28 |
ENSRNOT00000089538
|
Nf1
|
neurofibromin 1 |
| chr16_+_20740826 | 1.25 |
ENSRNOT00000038057
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chr7_+_59349745 | 1.25 |
ENSRNOT00000085334
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr3_+_148510779 | 1.22 |
ENSRNOT00000012156
|
Xkr7
|
XK related 7 |
| chr3_-_91370167 | 1.21 |
ENSRNOT00000006291
|
Prr5l
|
proline rich 5 like |
| chr9_+_62291809 | 1.21 |
ENSRNOT00000090621
|
Plcl1
|
phospholipase C-like 1 |
| chr5_+_90338795 | 1.20 |
ENSRNOT00000077864
ENSRNOT00000058882 |
LOC298139
|
similar to RIKEN cDNA 2310003M01 |
| chr8_+_104040934 | 1.17 |
ENSRNOT00000081204
|
Tfdp2
|
transcription factor Dp-2 |
| chr2_+_102685513 | 1.13 |
ENSRNOT00000033940
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr4_+_157524423 | 1.12 |
ENSRNOT00000036654
|
Zfp384
|
zinc finger protein 384 |
| chr13_+_95589668 | 1.11 |
ENSRNOT00000005849
|
Zfp238
|
zinc finger protein 238 |
| chr8_+_128824508 | 1.11 |
ENSRNOT00000025343
|
Mobp
|
myelin-associated oligodendrocyte basic protein |
| chr4_-_82194927 | 1.10 |
ENSRNOT00000072302
|
LOC103692128
|
homeobox protein Hox-A9 |
| chr4_-_87111025 | 1.07 |
ENSRNOT00000018442
|
Kbtbd2
|
kelch repeat and BTB domain containing 2 |
| chr1_+_142679345 | 1.07 |
ENSRNOT00000034267
|
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr6_-_27024129 | 1.06 |
ENSRNOT00000012273
|
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr7_+_72772440 | 0.98 |
ENSRNOT00000009989
|
Mtdh
|
metadherin |
| chr1_+_12823363 | 0.95 |
ENSRNOT00000086790
|
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr9_-_43139870 | 0.94 |
ENSRNOT00000021956
|
Sema4c
|
semaphorin 4C |
| chr18_+_27558089 | 0.93 |
ENSRNOT00000027499
|
Fam53c
|
family with sequence similarity 53, member C |
| chr13_-_76049363 | 0.93 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr2_+_69415057 | 0.92 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr6_+_29061618 | 0.91 |
ENSRNOT00000092223
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr18_+_47740328 | 0.89 |
ENSRNOT00000025119
|
Sncaip
|
synuclein, alpha interacting protein |
| chr6_+_83421882 | 0.88 |
ENSRNOT00000084258
|
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr8_-_46757781 | 0.88 |
ENSRNOT00000085294
|
AABR07073392.1
|
|
| chr9_+_40817654 | 0.88 |
ENSRNOT00000037392
|
AABR07067339.1
|
|
| chr10_+_35872619 | 0.84 |
ENSRNOT00000059190
|
Hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr10_+_72909550 | 0.83 |
ENSRNOT00000004540
|
Ppm1d
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
| chr15_-_25505691 | 0.78 |
ENSRNOT00000074488
|
LOC100911492
|
homeobox protein OTX2-like |
| chr14_+_23166633 | 0.73 |
ENSRNOT00000060109
|
Tmprss11g
|
transmembrane protease, serine 11G |
| chr7_-_104541392 | 0.71 |
ENSRNOT00000078116
|
Fam49b
|
family with sequence similarity 49, member B |
| chr20_-_43932361 | 0.71 |
ENSRNOT00000091030
|
Rfpl4b
|
ret finger protein-like 4B |
| chr2_+_189865915 | 0.69 |
ENSRNOT00000077362
|
Ilf2
|
interleukin enhancer binding factor 2 |
| chr8_-_110813000 | 0.63 |
ENSRNOT00000010634
|
Ephb1
|
Eph receptor B1 |
| chr13_-_50761306 | 0.63 |
ENSRNOT00000076610
ENSRNOT00000004241 |
Prelp
|
proline and arginine rich end leucine rich repeat protein |
| chr13_+_90533365 | 0.56 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr9_-_23493081 | 0.54 |
ENSRNOT00000072144
|
Rhag
|
Rh-associated glycoprotein |
| chr3_+_14990652 | 0.52 |
ENSRNOT00000090735
|
Dab2ip
|
DAB2 interacting protein |
| chr14_+_8080275 | 0.51 |
ENSRNOT00000065965
ENSRNOT00000092542 |
Mapk10
|
mitogen activated protein kinase 10 |
| chr14_-_41786084 | 0.50 |
ENSRNOT00000059439
|
Grxcr1
|
glutaredoxin and cysteine rich domain containing 1 |
| chr4_+_7122890 | 0.50 |
ENSRNOT00000076339
ENSRNOT00000076418 ENSRNOT00000014718 |
Abcf2
|
ATP binding cassette subfamily F member 2 |
| chr6_-_105518725 | 0.50 |
ENSRNOT00000009693
ENSRNOT00000083488 |
Map3k9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr1_+_29191192 | 0.48 |
ENSRNOT00000018718
|
Hey2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr10_-_87578854 | 0.48 |
ENSRNOT00000065619
|
LOC680160
|
similar to keratin associated protein 4-7 |
| chrX_-_16929907 | 0.47 |
ENSRNOT00000003971
|
Shroom4
|
shroom family member 4 |
| chr2_-_95106157 | 0.44 |
ENSRNOT00000041334
|
LOC499573
|
LRRGT00056 |
| chr6_-_99214251 | 0.41 |
ENSRNOT00000042682
|
Esr2
|
estrogen receptor 2 |
| chr20_-_5140304 | 0.37 |
ENSRNOT00000092646
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr11_-_84633504 | 0.37 |
ENSRNOT00000052120
|
Klhl24
|
kelch-like family member 24 |
| chr4_+_118207862 | 0.36 |
ENSRNOT00000085787
ENSRNOT00000023103 |
Tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr1_-_156327352 | 0.36 |
ENSRNOT00000074282
|
LOC100911776
|
coiled-coil domain-containing protein 89-like |
| chr14_+_8080565 | 0.36 |
ENSRNOT00000092395
|
Mapk10
|
mitogen activated protein kinase 10 |
| chr1_+_274030978 | 0.35 |
ENSRNOT00000076387
|
Mxi1
|
MAX interactor 1, dimerization protein |
| chr14_-_45376127 | 0.34 |
ENSRNOT00000059247
|
AABR07015015.2
|
|
| chr5_-_93244202 | 0.33 |
ENSRNOT00000075474
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr8_+_79489790 | 0.30 |
ENSRNOT00000091722
|
Prtg
|
protogenin |
| chr19_-_25345790 | 0.29 |
ENSRNOT00000010050
|
Zswim4
|
zinc finger, SWIM-type containing 4 |
| chr3_+_57286892 | 0.29 |
ENSRNOT00000039065
|
LOC103690018
|
Golgi reassembly-stacking protein 2-like |
| chrX_-_70460536 | 0.29 |
ENSRNOT00000076824
|
Pdzd11
|
PDZ domain containing 11 |
| chr2_-_96509424 | 0.28 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chrX_-_81866370 | 0.27 |
ENSRNOT00000042570
|
AABR07039651.1
|
|
| chr3_-_94418711 | 0.26 |
ENSRNOT00000089554
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr9_+_121456623 | 0.26 |
ENSRNOT00000056255
|
Ppidl1
|
peptidylprolyl isomerase D-like 1 |
| chrX_+_63343546 | 0.25 |
ENSRNOT00000076315
|
Klhl15
|
kelch-like family member 15 |
| chr13_+_76942928 | 0.25 |
ENSRNOT00000040505
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr1_+_259926537 | 0.22 |
ENSRNOT00000073537
|
NEWGENE_1306399
|
cyclin J |
| chr2_-_197935567 | 0.21 |
ENSRNOT00000085404
|
Rprd2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr1_+_240908483 | 0.20 |
ENSRNOT00000019367
|
Klf9
|
Kruppel-like factor 9 |
| chrX_-_73778595 | 0.19 |
ENSRNOT00000076081
ENSRNOT00000075926 ENSRNOT00000003782 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr13_+_76943633 | 0.18 |
ENSRNOT00000076908
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr1_+_260798239 | 0.12 |
ENSRNOT00000036791
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr3_+_80614937 | 0.09 |
ENSRNOT00000065462
|
Harbi1
|
harbinger transposase derived 1 |
| chr14_-_51462721 | 0.09 |
ENSRNOT00000015044
|
Hmgb3
|
high mobility group box 3 |
| chr7_-_129724303 | 0.04 |
ENSRNOT00000006034
|
Brd1
|
bromodomain containing 1 |
| chr16_-_71203609 | 0.04 |
ENSRNOT00000088458
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr13_+_47380406 | 0.04 |
ENSRNOT00000050289
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr3_-_94833406 | 0.03 |
ENSRNOT00000049695
|
RGD1560017
|
similar to Ac2-210 |
| chr1_+_154606490 | 0.02 |
ENSRNOT00000024095
|
Ccdc89
|
coiled-coil domain containing 89 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox5_Sry
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.4 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 3.5 | 10.5 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) positive regulation of metalloendopeptidase activity(GO:1904685) |
| 3.3 | 13.3 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 3.0 | 26.8 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 1.9 | 19.2 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 1.8 | 12.9 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 1.6 | 8.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 1.2 | 10.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.1 | 4.4 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.1 | 9.9 | GO:1904116 | response to vasopressin(GO:1904116) |
| 1.0 | 10.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.0 | 9.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.0 | 6.1 | GO:1905050 | positive regulation of metallopeptidase activity(GO:1905050) |
| 1.0 | 2.9 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.9 | 3.6 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.9 | 6.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.8 | 7.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.8 | 6.1 | GO:0071000 | response to magnetism(GO:0071000) |
| 0.5 | 29.7 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.5 | 2.1 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.5 | 4.0 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.5 | 5.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.4 | 2.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.4 | 2.2 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.4 | 3.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.4 | 1.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.4 | 8.8 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.4 | 9.7 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.4 | 4.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.4 | 1.5 | GO:0021586 | pons maturation(GO:0021586) |
| 0.4 | 2.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 3.4 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.3 | 3.3 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.3 | 3.9 | GO:0046549 | retinal rod cell development(GO:0046548) retinal cone cell development(GO:0046549) |
| 0.3 | 1.9 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.3 | 1.3 | GO:0061534 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.3 | 2.9 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.3 | 1.4 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.3 | 2.2 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.3 | 1.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.3 | 2.9 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.3 | 3.8 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 17.9 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.2 | 0.7 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.2 | 1.0 | GO:0061428 | embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.2 | 2.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 1.4 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.2 | 6.2 | GO:0097435 | definitive hemopoiesis(GO:0060216) fibril organization(GO:0097435) |
| 0.2 | 2.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.2 | 5.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 1.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 3.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.2 | 2.7 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 2.4 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 | 2.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.3 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 1.7 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 3.4 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 2.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 1.4 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 1.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 2.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 1.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 14.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 0.9 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.9 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.1 | 0.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.8 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 4.0 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.1 | 2.7 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.4 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 1.5 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.1 | 1.4 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 3.2 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.1 | 0.4 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 1.6 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.9 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 3.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 3.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 5.5 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 3.8 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.8 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 3.4 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 1.0 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.0 | 1.4 | GO:0034698 | response to gonadotropin(GO:0034698) |
| 0.0 | 6.2 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 0.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.1 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.5 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.7 | 10.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.3 | 10.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.1 | 34.1 | GO:0031430 | M band(GO:0031430) |
| 0.9 | 10.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.6 | 6.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.5 | 6.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.5 | 4.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.5 | 1.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.5 | 2.8 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.4 | 6.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 2.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 1.6 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.3 | 2.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 16.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 3.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.3 | 5.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 9.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 6.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 0.5 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.2 | 2.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 2.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 3.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 7.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 34.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 10.7 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 1.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 5.9 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.1 | 1.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 2.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 16.4 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 41.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 8.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 2.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 7.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 2.3 | 13.5 | GO:0098821 | activin receptor activity, type I(GO:0016361) BMP receptor activity(GO:0098821) |
| 2.0 | 9.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.6 | 29.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.5 | 9.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 1.3 | 16.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.0 | 11.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.9 | 7.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.8 | 3.9 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.7 | 2.9 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.7 | 5.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.6 | 1.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.6 | 1.7 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.6 | 4.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 12.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.5 | 2.9 | GO:0035473 | lipase binding(GO:0035473) |
| 0.4 | 4.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 1.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 3.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 2.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 0.7 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.2 | 9.0 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.2 | 12.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 1.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 2.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.2 | 5.0 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 1.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 0.5 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 1.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 4.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 2.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.4 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 6.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 8.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.5 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 2.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 8.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 4.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 4.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 1.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 3.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 2.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 4.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 7.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 3.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 8.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 2.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 12.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 5.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 29.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 13.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.4 | 9.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.4 | 6.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.4 | 16.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 14.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 10.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 6.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 5.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 22.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 22.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 10.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 5.6 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 1.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 27.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 5.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.4 | 9.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.4 | 11.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 4.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.3 | 13.7 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.3 | 2.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 3.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 4.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 3.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 2.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.7 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 1.4 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 2.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 3.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.4 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.1 | 7.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 4.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 2.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |