Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
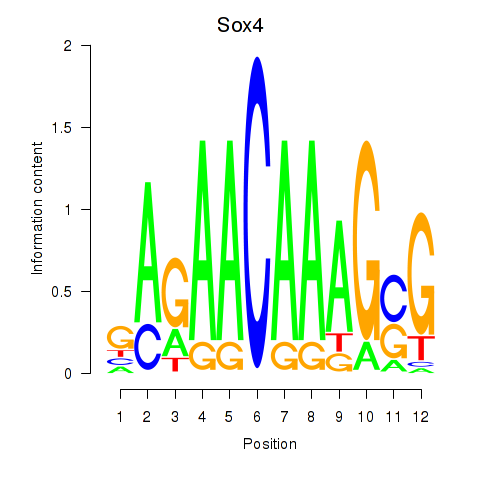
Results for Sox4
Z-value: 0.56
Transcription factors associated with Sox4
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Sox4 motif
Sorted Z-values of Sox4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_139528162 | 14.78 |
ENSRNOT00000014317
|
Slc7a11
|
solute carrier family 7 member 11 |
| chr18_+_79406381 | 13.91 |
ENSRNOT00000022303
ENSRNOT00000058295 ENSRNOT00000058296 ENSRNOT00000022280 |
Mbp
|
myelin basic protein |
| chr9_-_100253609 | 13.12 |
ENSRNOT00000036061
|
Kif1a
|
kinesin family member 1A |
| chr8_+_128824508 | 13.11 |
ENSRNOT00000025343
|
Mobp
|
myelin-associated oligodendrocyte basic protein |
| chr1_+_234363994 | 13.04 |
ENSRNOT00000018137
|
Rorb
|
RAR-related orphan receptor B |
| chr9_+_117044533 | 12.52 |
ENSRNOT00000072916
|
Tmem200c
|
transmembrane protein 200C |
| chr3_-_91217491 | 11.87 |
ENSRNOT00000006115
|
Rag1
|
recombination activating 1 |
| chrX_-_118513061 | 10.94 |
ENSRNOT00000043338
|
Il13ra2
|
interleukin 13 receptor subunit alpha 2 |
| chr6_-_94980004 | 10.45 |
ENSRNOT00000006373
|
Rtn1
|
reticulon 1 |
| chr10_+_57040267 | 9.88 |
ENSRNOT00000026207
|
Arrb2
|
arrestin, beta 2 |
| chr13_+_83996080 | 9.77 |
ENSRNOT00000004403
ENSRNOT00000070958 |
Cd247
|
Cd247 molecule |
| chrX_+_65566047 | 9.67 |
ENSRNOT00000092103
|
Heph
|
hephaestin |
| chr14_-_44845218 | 7.68 |
ENSRNOT00000004003
|
Klhl5
|
kelch-like family member 5 |
| chr1_+_101214593 | 7.44 |
ENSRNOT00000028086
|
Tead2
|
TEA domain transcription factor 2 |
| chr15_+_18399733 | 7.33 |
ENSRNOT00000061158
|
Fam107a
|
family with sequence similarity 107, member A |
| chr5_+_121952977 | 6.69 |
ENSRNOT00000008278
|
Pde4b
|
phosphodiesterase 4B |
| chr5_+_147714163 | 6.62 |
ENSRNOT00000012663
|
Marcksl1
|
MARCKS-like 1 |
| chr13_+_51972974 | 6.31 |
ENSRNOT00000008164
|
Arl8a
|
ADP-ribosylation factor like GTPase 8A |
| chr6_-_92527711 | 5.98 |
ENSRNOT00000007529
|
Nin
|
ninein |
| chr3_+_72134731 | 5.92 |
ENSRNOT00000083592
|
Ypel4
|
yippee-like 4 |
| chr3_+_149144795 | 5.73 |
ENSRNOT00000015482
|
Dnmt3b
|
DNA methyltransferase 3 beta |
| chr3_+_162357356 | 5.41 |
ENSRNOT00000030119
|
Eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr1_-_48825364 | 5.37 |
ENSRNOT00000024213
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chrX_-_159891326 | 5.26 |
ENSRNOT00000001154
|
Rbmx
|
RNA binding motif protein, X-linked |
| chr15_+_18399515 | 5.15 |
ENSRNOT00000071273
|
Fam107a
|
family with sequence similarity 107, member A |
| chr10_+_56453877 | 5.05 |
ENSRNOT00000031640
|
Plscr3
|
phospholipid scramblase 3 |
| chr19_+_46733633 | 4.82 |
ENSRNOT00000016307
|
Clec3a
|
C-type lectin domain family 3, member A |
| chr8_+_2604962 | 4.72 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chr16_+_83206004 | 4.68 |
ENSRNOT00000018870
|
Ankrd10
|
ankyrin repeat domain 10 |
| chr20_+_3558827 | 4.65 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr5_-_147953093 | 4.28 |
ENSRNOT00000075270
|
Khdrbs1
|
KH RNA binding domain containing, signal transduction associated 1 |
| chr15_+_33600102 | 4.24 |
ENSRNOT00000022664
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr2_-_236431596 | 4.10 |
ENSRNOT00000055570
ENSRNOT00000089674 |
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chrX_+_18489597 | 3.95 |
ENSRNOT00000059744
|
LOC681355
|
similar to potassium channel tetramerisation domain containing 12b |
| chr10_+_110445797 | 3.65 |
ENSRNOT00000054920
|
Narf
|
nuclear prelamin A recognition factor |
| chr6_-_18351536 | 3.56 |
ENSRNOT00000061588
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr6_+_44009872 | 3.32 |
ENSRNOT00000082657
|
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr10_-_83655182 | 3.18 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr20_+_13670066 | 3.07 |
ENSRNOT00000031400
|
LOC103694874
|
stromelysin-3 |
| chr4_+_13405136 | 2.95 |
ENSRNOT00000091004
|
Gnai1
|
G protein subunit alpha i1 |
| chr15_+_33600337 | 2.92 |
ENSRNOT00000075965
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr5_+_101526551 | 2.86 |
ENSRNOT00000015254
|
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr7_-_59514939 | 2.73 |
ENSRNOT00000085579
|
Kcnmb4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr1_-_170628915 | 2.67 |
ENSRNOT00000042865
|
Dchs1
|
dachsous cadherin-related 1 |
| chr3_-_118051289 | 2.52 |
ENSRNOT00000011551
|
Secisbp2l
|
SECIS binding protein 2-like |
| chr15_-_28672530 | 2.30 |
ENSRNOT00000022593
ENSRNOT00000087722 |
Chd8
|
chromodomain helicase DNA binding protein 8 |
| chr16_-_69961162 | 2.26 |
ENSRNOT00000074955
|
Prrg3
|
proline rich and Gla domain 3 |
| chr17_+_42168263 | 2.26 |
ENSRNOT00000047422
|
LOC100360654
|
ribosomal protein L37-like |
| chr4_-_77347011 | 2.24 |
ENSRNOT00000008149
|
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr19_+_755460 | 2.23 |
ENSRNOT00000076560
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chr5_+_148781222 | 2.19 |
ENSRNOT00000015831
|
Pum1
|
pumilio RNA-binding family member 1 |
| chr11_-_31213787 | 2.19 |
ENSRNOT00000002800
|
Paxbp1
|
PAX3 and PAX7 binding protein 1 |
| chr1_-_170594168 | 2.15 |
ENSRNOT00000026280
|
Tpp1
|
tripeptidyl peptidase 1 |
| chr18_+_30909490 | 2.15 |
ENSRNOT00000026967
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8 |
| chr9_+_46657575 | 1.91 |
ENSRNOT00000083322
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr3_-_5976244 | 1.75 |
ENSRNOT00000009893
|
Vav2
|
vav guanine nucleotide exchange factor 2 |
| chr1_-_169005190 | 1.70 |
ENSRNOT00000043719
|
Hbe1
|
hemoglobin subunit epsilon 1 |
| chr12_+_23789638 | 1.68 |
ENSRNOT00000001954
ENSRNOT00000084730 |
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma |
| chr6_+_108820774 | 1.67 |
ENSRNOT00000089683
ENSRNOT00000080038 ENSRNOT00000006492 |
Ylpm1
|
YLP motif containing 1 |
| chr6_+_106052212 | 1.63 |
ENSRNOT00000010626
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chrX_-_14220662 | 1.38 |
ENSRNOT00000045753
|
Srpx
|
sushi-repeat-containing protein, X-linked |
| chr2_-_250235435 | 1.34 |
ENSRNOT00000088618
|
Lmo4
|
LIM domain only 4 |
| chr3_+_95715193 | 1.17 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr8_+_69971778 | 1.13 |
ENSRNOT00000058007
ENSRNOT00000037941 ENSRNOT00000050649 |
Megf11
|
multiple EGF-like-domains 11 |
| chr3_-_3541947 | 1.11 |
ENSRNOT00000024786
|
Nacc2
|
NACC family member 2 |
| chr10_-_90415070 | 1.04 |
ENSRNOT00000055179
|
Itga2b
|
integrin subunit alpha 2b |
| chr7_-_12405022 | 0.98 |
ENSRNOT00000021648
|
Cirbp
|
cold inducible RNA binding protein |
| chr7_-_60416925 | 0.67 |
ENSRNOT00000008193
|
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr7_-_98197087 | 0.48 |
ENSRNOT00000010484
ENSRNOT00000079961 |
Klhl38
|
kelch-like family member 38 |
| chr18_+_60392376 | 0.35 |
ENSRNOT00000023890
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr1_+_264670841 | 0.33 |
ENSRNOT00000034814
ENSRNOT00000082412 |
Slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr18_+_15467870 | 0.28 |
ENSRNOT00000091696
|
B4galt6
|
beta-1,4-galactosyltransferase 6 |
| chr10_+_75365822 | 0.09 |
ENSRNOT00000055705
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chr1_+_190671696 | 0.06 |
ENSRNOT00000084934
|
AABR07005633.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.9 | GO:1904685 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) positive regulation of metalloendopeptidase activity(GO:1904685) |
| 3.0 | 11.9 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 2.5 | 9.9 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 1.9 | 5.7 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 1.4 | 4.3 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 1.2 | 4.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 1.1 | 6.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 1.1 | 10.9 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 1.1 | 13.0 | GO:0046549 | amacrine cell differentiation(GO:0035881) retinal rod cell development(GO:0046548) retinal cone cell development(GO:0046549) |
| 0.9 | 2.7 | GO:0003192 | mitral valve formation(GO:0003192) condensed mesenchymal cell proliferation(GO:0072137) |
| 0.9 | 14.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.7 | 6.0 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.7 | 3.6 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.6 | 5.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.6 | 5.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.5 | 4.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.4 | 1.2 | GO:0003322 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.4 | 2.2 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.4 | 7.4 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.3 | 5.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.3 | 2.5 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.3 | 2.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.2 | 7.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 2.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 | 2.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.2 | 1.4 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 9.8 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.2 | 3.0 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.2 | 4.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 5.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 2.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 3.1 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 2.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 1.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 1.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.3 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 1.0 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 1.9 | GO:0070571 | negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 1.3 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.1 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 9.8 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.1 | 2.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.7 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 1.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 3.9 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 4.7 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 12.5 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 1.7 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 1.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 3.3 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.3 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.7 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 3.2 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 2.3 | 13.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.2 | 9.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.1 | 5.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.9 | 4.7 | GO:0097169 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.9 | 6.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.5 | 3.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.3 | 6.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 9.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 6.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 13.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 1.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 9.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 14.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 2.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 5.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 2.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 3.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 5.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 3.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 14.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 14.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 11.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 3.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 4.3 | GO:0098797 | plasma membrane protein complex(GO:0098797) |
| 0.0 | 3.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 3.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 10.9 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.8 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 3.3 | 9.9 | GO:0031826 | follicle-stimulating hormone receptor binding(GO:0031762) type 2A serotonin receptor binding(GO:0031826) |
| 2.6 | 13.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 2.3 | 27.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.1 | 5.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.8 | 4.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.7 | 3.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.7 | 4.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.6 | 1.7 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.5 | 2.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.4 | 5.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.4 | 2.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.4 | 7.4 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.4 | 6.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.4 | 4.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.3 | 2.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 4.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.3 | 1.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 2.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 5.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 2.2 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.2 | 1.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.2 | 3.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 13.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.0 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 11.4 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 1.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 4.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 2.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 11.9 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.1 | 5.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.3 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 7.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 1.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 5.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 5.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 3.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 4.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 10.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 3.3 | GO:0016746 | transferase activity, transferring acyl groups(GO:0016746) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 10.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.5 | 9.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.3 | 9.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 4.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 3.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.9 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 14.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.4 | 4.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 9.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 7.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 3.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 6.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 13.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 5.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 3.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 4.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 4.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 2.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 5.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 2.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |