Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
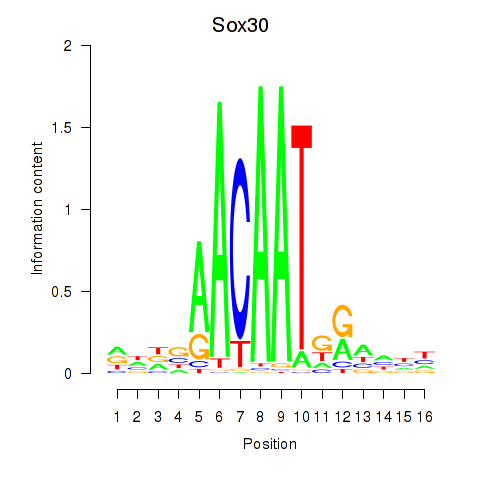
Results for Sox30
Z-value: 0.69
Transcription factors associated with Sox30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox30
|
ENSRNOG00000006098 | SRY box 30 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox30 | rn6_v1_chr10_+_31074251_31074251 | -0.03 | 6.2e-01 | Click! |
Activity profile of Sox30 motif
Sorted Z-values of Sox30 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_44086006 | 37.50 |
ENSRNOT00000034449
ENSRNOT00000082604 |
Ermn
|
ermin |
| chr1_+_12915734 | 37.48 |
ENSRNOT00000089066
|
Txlnb
|
taxilin beta |
| chr5_+_64326733 | 24.35 |
ENSRNOT00000065775
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr5_+_90338795 | 22.62 |
ENSRNOT00000077864
ENSRNOT00000058882 |
LOC298139
|
similar to RIKEN cDNA 2310003M01 |
| chr4_-_11610518 | 18.18 |
ENSRNOT00000066643
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr5_-_133959447 | 17.41 |
ENSRNOT00000011985
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr9_+_73418607 | 16.77 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr10_+_82775691 | 15.74 |
ENSRNOT00000030737
|
Hils1
|
histone linker H1 domain, spermatid-specific 1 |
| chr4_-_39102807 | 15.16 |
ENSRNOT00000052063
|
Thsd7a
|
thrombospondin type 1 domain containing 7A |
| chr13_+_87986240 | 14.99 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr13_+_57243877 | 14.82 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr14_-_84106997 | 14.70 |
ENSRNOT00000065501
|
Osbp2
|
oxysterol binding protein 2 |
| chr10_+_53778662 | 14.44 |
ENSRNOT00000045718
|
Myh2
|
myosin heavy chain 2 |
| chr7_+_130042947 | 12.55 |
ENSRNOT00000089707
|
Panx2
|
pannexin 2 |
| chr6_-_137084739 | 12.24 |
ENSRNOT00000060943
|
Tmem179
|
transmembrane protein 179 |
| chr3_-_72219246 | 11.33 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chrX_+_62727755 | 11.11 |
ENSRNOT00000077055
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr4_-_166803127 | 10.58 |
ENSRNOT00000067909
|
RGD1306750
|
LOC362451 |
| chr18_+_30826260 | 9.95 |
ENSRNOT00000065235
|
Pcdhgb1
|
protocadherin gamma subfamily B, 1 |
| chr8_-_62987182 | 9.67 |
ENSRNOT00000070885
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chrM_+_3904 | 9.18 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr2_-_117454769 | 8.16 |
ENSRNOT00000068381
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr18_+_28781349 | 7.80 |
ENSRNOT00000026112
|
Psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr8_-_90664554 | 6.57 |
ENSRNOT00000039917
ENSRNOT00000074896 ENSRNOT00000072225 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr4_-_51199570 | 6.54 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr6_-_51019407 | 6.47 |
ENSRNOT00000011659
|
Gpr22
|
G protein-coupled receptor 22 |
| chr7_-_107223047 | 6.16 |
ENSRNOT00000007250
ENSRNOT00000084875 |
Lrrc6
|
leucine rich repeat containing 6 |
| chr5_+_152606847 | 5.61 |
ENSRNOT00000089070
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr18_+_30875011 | 5.28 |
ENSRNOT00000027009
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr18_+_30890869 | 4.98 |
ENSRNOT00000060466
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr2_+_188087486 | 4.41 |
ENSRNOT00000027455
|
Rit1
|
Ras-like without CAAX 1 |
| chr10_-_11005952 | 3.82 |
ENSRNOT00000005031
|
Hmox2
|
heme oxygenase 2 |
| chr2_+_189865915 | 3.69 |
ENSRNOT00000077362
|
Ilf2
|
interleukin enhancer binding factor 2 |
| chr2_+_93520734 | 3.68 |
ENSRNOT00000083909
|
Snx16
|
sorting nexin 16 |
| chrX_+_70596576 | 3.68 |
ENSRNOT00000045082
ENSRNOT00000003741 ENSRNOT00000076079 |
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr6_-_41039437 | 3.67 |
ENSRNOT00000005774
|
Trib2
|
tribbles pseudokinase 2 |
| chr1_-_171009670 | 2.70 |
ENSRNOT00000026487
|
Olr222
|
olfactory receptor 222 |
| chr1_+_13595295 | 2.06 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr3_-_76250840 | 1.34 |
ENSRNOT00000007782
|
Olr607
|
olfactory receptor 607 |
| chr4_-_165747201 | 1.01 |
ENSRNOT00000007435
|
Tas2r130
|
taste receptor, type 2, member 130 |
| chr7_-_9136515 | 0.57 |
ENSRNOT00000081964
|
Olr1059
|
olfactory receptor 1059 |
| chr11_-_70585053 | 0.47 |
ENSRNOT00000086955
|
Zfp148
|
zinc finger protein 148 |
| chr6_-_104290579 | 0.44 |
ENSRNOT00000066014
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr14_-_81399353 | 0.30 |
ENSRNOT00000018340
|
Add1
|
adducin 1 |
| chr7_+_4620033 | 0.16 |
ENSRNOT00000072277
|
AABR07055511.1
|
|
| chr5_-_157059508 | 0.15 |
ENSRNOT00000022282
|
Vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr16_+_74237001 | 0.08 |
ENSRNOT00000026039
|
Polb
|
DNA polymerase beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox30
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 18.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 1.4 | 12.5 | GO:0034214 | protein hexamerization(GO:0034214) |
| 1.3 | 3.8 | GO:0006788 | heme oxidation(GO:0006788) |
| 1.2 | 11.1 | GO:1904116 | response to vasopressin(GO:1904116) |
| 1.1 | 14.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.9 | 3.7 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.9 | 8.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.7 | 6.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.4 | 15.7 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.4 | 16.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.4 | 9.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 6.5 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.3 | 3.7 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.3 | 7.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 15.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.2 | 37.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.2 | 3.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 8.0 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 9.7 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 22.6 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 6.6 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 14.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 14.8 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 0.3 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 5.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 4.4 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 2.4 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 14.3 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.0 | 6.5 | GO:1901652 | response to peptide(GO:1901652) |
| 0.0 | 0.5 | GO:0021762 | substantia nigra development(GO:0021762) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 37.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.9 | 16.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.8 | 18.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 1.6 | 14.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.7 | 15.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.5 | 14.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.4 | 3.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 12.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.4 | 11.3 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 9.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 14.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 3.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 8.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 17.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 18.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 6.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 22.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 6.2 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 4.5 | GO:0030425 | dendrite(GO:0030425) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 18.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 3.1 | 12.5 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 2.2 | 11.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.3 | 3.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.9 | 5.6 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.7 | 16.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.6 | 11.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 14.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.4 | 17.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.4 | 6.5 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.4 | 3.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 9.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 14.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.3 | 37.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.3 | 7.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 37.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 22.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 6.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 3.7 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 15.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 14.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 3.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 7.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 3.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 4.4 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 14.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 16.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 4.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 18.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.5 | 6.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 11.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 3.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 4.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 3.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 12.5 | REACTOME NEURONAL SYSTEM | Genes involved in Neuronal System |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |