Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
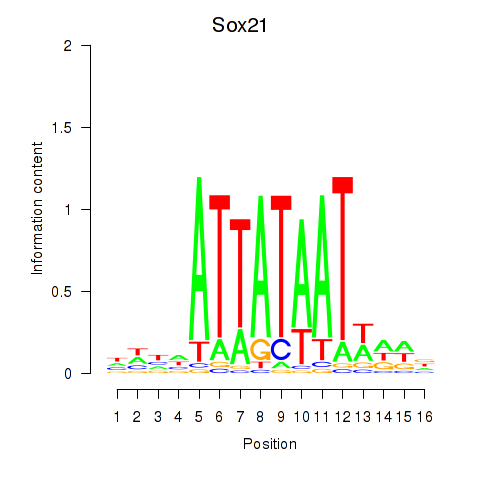
Results for Sox21
Z-value: 1.31
Transcription factors associated with Sox21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox21
|
ENSRNOG00000052579 | Sox21 |
Activity profile of Sox21 motif
Sorted Z-values of Sox21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_17889972 | 96.28 |
ENSRNOT00000073021
|
AABR07051611.1
|
|
| chr8_-_114617466 | 73.44 |
ENSRNOT00000082992
ENSRNOT00000038160 |
Col6a5
|
collagen type VI alpha 5 chain |
| chr2_-_173668555 | 59.00 |
ENSRNOT00000013452
|
Serpini2
|
serpin family I member 2 |
| chr4_+_14109864 | 57.84 |
ENSRNOT00000076349
|
RGD1565355
|
similar to fatty acid translocase/CD36 |
| chr6_-_138536321 | 56.59 |
ENSRNOT00000077743
|
AABR07065643.1
|
|
| chr6_-_138249382 | 52.78 |
ENSRNOT00000085678
ENSRNOT00000006912 |
Ighm
|
immunoglobulin heavy constant mu |
| chr6_+_139177200 | 48.45 |
ENSRNOT00000084131
|
AABR07065676.1
|
|
| chr6_-_140715174 | 47.46 |
ENSRNOT00000085345
|
AABR07065773.1
|
|
| chr6_+_139209936 | 43.06 |
ENSRNOT00000087620
|
AABR07065680.1
|
|
| chr3_+_16846412 | 40.82 |
ENSRNOT00000074266
|
AABR07051551.1
|
|
| chr4_+_98481520 | 38.04 |
ENSRNOT00000078381
ENSRNOT00000048493 |
AABR07060886.1
|
|
| chr1_+_22332090 | 37.32 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr3_+_17546566 | 36.90 |
ENSRNOT00000050825
|
AABR07051583.1
|
|
| chr9_+_8054466 | 36.23 |
ENSRNOT00000081513
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr7_-_14418950 | 34.73 |
ENSRNOT00000066722
|
Rasal3
|
RAS protein activator like 3 |
| chr5_+_157222636 | 32.93 |
ENSRNOT00000022579
|
Pla2g2d
|
phospholipase A2, group IID |
| chr6_+_139345486 | 32.81 |
ENSRNOT00000081540
|
AABR07065688.1
|
|
| chr13_+_92264231 | 32.49 |
ENSRNOT00000066509
ENSRNOT00000004716 |
Spta1
|
spectrin, alpha, erythrocytic 1 |
| chr3_+_20007192 | 32.39 |
ENSRNOT00000075229
|
AABR07051716.1
|
|
| chr6_-_138550576 | 30.61 |
ENSRNOT00000075284
|
AABR07065645.1
|
|
| chr4_-_163762434 | 30.09 |
ENSRNOT00000081854
|
Ly49si1
|
immunoreceptor Ly49si1 |
| chr8_+_5768811 | 29.42 |
ENSRNOT00000013936
|
Mmp8
|
matrix metallopeptidase 8 |
| chr6_-_138536162 | 29.18 |
ENSRNOT00000083031
|
AABR07065643.1
|
|
| chr6_-_139654508 | 26.65 |
ENSRNOT00000082576
|
AABR07065705.5
|
|
| chr6_-_138909105 | 25.89 |
ENSRNOT00000087855
|
AABR07065656.9
|
|
| chr6_-_138948583 | 25.85 |
ENSRNOT00000084614
|
AABR07065656.2
|
|
| chr14_+_108826831 | 25.37 |
ENSRNOT00000083146
ENSRNOT00000009421 |
Bcl11a
|
B-cell CLL/lymphoma 11A |
| chr6_-_138852571 | 24.72 |
ENSRNOT00000081803
|
AABR07065656.8
|
|
| chr17_-_43776460 | 23.67 |
ENSRNOT00000089055
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr19_-_37796089 | 23.57 |
ENSRNOT00000024891
|
Ranbp10
|
RAN binding protein 10 |
| chr6_-_138679665 | 22.31 |
ENSRNOT00000086777
|
AABR07065651.4
|
|
| chr6_-_142372031 | 21.88 |
ENSRNOT00000063929
|
AABR07065814.3
|
|
| chr10_+_30038709 | 21.15 |
ENSRNOT00000005898
|
Il12b
|
interleukin 12B |
| chr1_+_242959488 | 19.88 |
ENSRNOT00000015668
|
Dock8
|
dedicator of cytokinesis 8 |
| chr17_-_86657473 | 19.45 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
| chr3_+_17139670 | 18.76 |
ENSRNOT00000073316
|
AABR07051564.1
|
|
| chr3_+_16590244 | 18.27 |
ENSRNOT00000073229
|
AABR07051535.1
|
|
| chr3_+_172550258 | 18.08 |
ENSRNOT00000075148
|
Tubb1
|
tubulin, beta 1 class VI |
| chrX_+_104684676 | 18.06 |
ENSRNOT00000083229
|
Tnmd
|
tenomodulin |
| chr20_-_45260119 | 18.02 |
ENSRNOT00000000718
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr1_+_253221812 | 17.20 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chr6_-_86914883 | 16.29 |
ENSRNOT00000091202
|
Mis18bp1
|
MIS18 binding protein 1 |
| chr20_-_4792283 | 16.10 |
ENSRNOT00000061027
|
RT1-CE1
|
RT1 class I, locus1 |
| chr4_+_93959152 | 16.02 |
ENSRNOT00000058437
|
AABR07060795.1
|
|
| chr5_-_12172009 | 14.43 |
ENSRNOT00000061903
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr8_+_48665652 | 13.75 |
ENSRNOT00000059715
|
H2afx
|
H2A histone family, member X |
| chr1_-_118826977 | 13.64 |
ENSRNOT00000079274
|
AABR07003833.1
|
|
| chr4_-_13878126 | 13.14 |
ENSRNOT00000007032
|
Gnat3
|
G protein subunit alpha transducin 3 |
| chr2_-_127710448 | 11.72 |
ENSRNOT00000093733
|
Mfsd8
|
major facilitator superfamily domain containing 8 |
| chr6_-_125723732 | 11.52 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr4_-_168517177 | 11.48 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
| chr6_+_135513650 | 11.27 |
ENSRNOT00000010676
|
Rcor1
|
REST corepressor 1 |
| chr4_-_164899041 | 11.08 |
ENSRNOT00000090316
|
Ly49i2
|
Ly49 inhibitory receptor 2 |
| chr1_+_75428027 | 10.41 |
ENSRNOT00000058334
|
Pla2g4c
|
phospholipase A2, group IVC |
| chr20_-_4792450 | 10.19 |
ENSRNOT00000091344
|
RT1-CE6
|
RT1 class I, locus CE6 |
| chr1_-_64090017 | 9.70 |
ENSRNOT00000086622
ENSRNOT00000091654 |
Rps9
|
ribosomal protein S9 |
| chr13_-_61306939 | 9.69 |
ENSRNOT00000086610
|
Rgs21
|
regulator of G-protein signaling 21 |
| chr4_+_153217782 | 9.11 |
ENSRNOT00000015499
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr1_-_53152866 | 9.04 |
ENSRNOT00000085501
|
Fgfr1op
|
Fgfr1 oncogene partner |
| chr15_+_80040842 | 8.71 |
ENSRNOT00000043065
|
RGD1306441
|
similar to RIKEN cDNA 4921530L21 |
| chr15_-_27968358 | 8.61 |
ENSRNOT00000040616
|
Rnase9
|
ribonuclease A family member 9 |
| chr8_-_104155775 | 8.38 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chr1_-_141504111 | 8.33 |
ENSRNOT00000040164
|
Snrpep2
|
small nuclear ribonucleoprotein polypeptide E pseudogene 2 |
| chr17_-_51912496 | 8.30 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr15_-_82581916 | 7.96 |
ENSRNOT00000057891
|
RGD1560069
|
similar to ribosomal protein L27 |
| chr2_-_248484100 | 7.91 |
ENSRNOT00000040856
|
Gbp3
|
guanylate binding protein 3 |
| chr10_-_35187416 | 7.82 |
ENSRNOT00000084509
|
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr3_-_111080705 | 7.57 |
ENSRNOT00000079339
|
Rhov
|
ras homolog family member V |
| chrX_-_10031167 | 7.54 |
ENSRNOT00000060988
|
Gpr34
|
G protein-coupled receptor 34 |
| chr2_-_118882562 | 7.48 |
ENSRNOT00000058860
|
Kcnmb3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr18_-_1828606 | 7.35 |
ENSRNOT00000033312
|
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr4_-_165118062 | 7.32 |
ENSRNOT00000087219
|
LOC502908
|
Ly49s8 |
| chr6_+_12179227 | 7.26 |
ENSRNOT00000061693
|
Foxn2
|
forkhead box N2 |
| chr20_+_4329811 | 6.97 |
ENSRNOT00000000513
|
Notch4
|
notch 4 |
| chr3_+_47439076 | 6.94 |
ENSRNOT00000011794
|
Tank
|
TRAF family member-associated NFKB activator |
| chr4_-_147122060 | 6.86 |
ENSRNOT00000047694
|
AC128901.1
|
|
| chr3_+_94585873 | 6.82 |
ENSRNOT00000076762
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr11_+_2645865 | 6.60 |
ENSRNOT00000000905
|
Pou1f1
|
POU class 1 homeobox 1 |
| chr13_-_91228901 | 6.54 |
ENSRNOT00000071728
ENSRNOT00000073643 ENSRNOT00000071897 |
LOC100911825
|
low affinity immunoglobulin gamma Fc region receptor III-like |
| chr7_-_104541392 | 6.54 |
ENSRNOT00000078116
|
Fam49b
|
family with sequence similarity 49, member B |
| chr3_-_132282879 | 6.51 |
ENSRNOT00000042040
|
LOC100361388
|
high-mobility group nucleosomal binding domain 2-like |
| chr10_-_1461216 | 6.47 |
ENSRNOT00000083125
ENSRNOT00000084447 ENSRNOT00000078421 |
Parn
|
poly(A)-specific ribonuclease |
| chr13_-_52413681 | 6.34 |
ENSRNOT00000037100
ENSRNOT00000011801 |
Nav1
|
neuron navigator 1 |
| chr10_-_103619083 | 6.23 |
ENSRNOT00000029092
|
Cd300e
|
Cd300e molecule |
| chr12_-_21450204 | 5.95 |
ENSRNOT00000065866
|
LOC100910636
|
zinc finger CW-type PWWP domain protein 1-like |
| chr5_-_120083904 | 5.79 |
ENSRNOT00000064542
|
Jak1
|
Janus kinase 1 |
| chr10_+_65552897 | 5.57 |
ENSRNOT00000056217
|
Spag5
|
sperm associated antigen 5 |
| chr3_+_22964230 | 5.50 |
ENSRNOT00000041813
|
LOC100362149
|
ribosomal protein S20-like |
| chr10_-_66873948 | 5.47 |
ENSRNOT00000039261
|
Evi2a
|
ecotropic viral integration site 2A |
| chr4_+_88026033 | 5.37 |
ENSRNOT00000074905
|
Vom1r80
|
vomeronasal 1 receptor 80 |
| chr1_-_219544328 | 5.33 |
ENSRNOT00000025847
|
Grk2
|
G protein-coupled receptor kinase 2 |
| chrY_-_1398030 | 5.31 |
ENSRNOT00000088719
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr15_-_57651041 | 5.09 |
ENSRNOT00000072138
|
Spert
|
spermatid associated |
| chr7_-_60122805 | 5.04 |
ENSRNOT00000037946
|
Cct2
|
chaperonin containing TCP1 subunit 2 |
| chr6_-_12997817 | 5.03 |
ENSRNOT00000022802
|
Fshr
|
follicle stimulating hormone receptor |
| chr1_-_102845083 | 5.03 |
ENSRNOT00000082289
|
Saa4
|
serum amyloid A4 |
| chr3_-_60023913 | 5.00 |
ENSRNOT00000064662
|
Cir1
|
corepressor interacting with RBPJ, 1 |
| chr4_+_136078391 | 4.93 |
ENSRNOT00000048672
|
AABR07061614.1
|
|
| chr1_+_102924059 | 4.82 |
ENSRNOT00000078137
|
AC099150.1
|
|
| chr3_+_98695503 | 4.79 |
ENSRNOT00000042764
|
AABR07053304.1
|
|
| chr7_+_38900593 | 4.69 |
ENSRNOT00000072166
|
Epyc
|
epiphycan |
| chr5_+_131915382 | 4.66 |
ENSRNOT00000087012
|
Skint1
|
selection and upkeep of intraepithelial T cells 1 |
| chr12_+_8725517 | 4.65 |
ENSRNOT00000001243
|
Slc46a3
|
solute carrier family 46, member 3 |
| chr1_-_172567846 | 4.62 |
ENSRNOT00000012974
|
Olr259
|
olfactory receptor 259 |
| chr10_-_87286387 | 4.62 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr13_+_110372164 | 4.48 |
ENSRNOT00000049634
|
RGD1560186
|
similar to ribosomal protein L37 |
| chr5_+_150787043 | 4.42 |
ENSRNOT00000082945
|
Eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr2_+_54466280 | 4.28 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr1_+_88955135 | 4.00 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chr11_+_47061354 | 3.99 |
ENSRNOT00000039997
|
Pcnp
|
PEST proteolytic signal containing nuclear protein |
| chr15_-_27140657 | 3.97 |
ENSRNOT00000016727
|
Olr1612
|
olfactory receptor 1612 |
| chrX_+_54062935 | 3.75 |
ENSRNOT00000004854
|
Tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr1_+_88955440 | 3.75 |
ENSRNOT00000091101
|
Prodh2
|
proline dehydrogenase 2 |
| chr5_+_121952977 | 3.72 |
ENSRNOT00000008278
|
Pde4b
|
phosphodiesterase 4B |
| chr16_+_50152008 | 3.71 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr3_+_43255567 | 3.69 |
ENSRNOT00000044419
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr4_-_119131202 | 3.68 |
ENSRNOT00000011675
|
Antxr1
|
anthrax toxin receptor 1 |
| chrX_-_122062799 | 3.53 |
ENSRNOT00000086852
|
Gm4907
|
predicted gene 4907 |
| chr1_-_256914260 | 3.51 |
ENSRNOT00000020620
|
Fra10ac1
|
FRA10A associated CGG repeat 1 |
| chr18_-_58081096 | 3.37 |
ENSRNOT00000059276
|
Spink13
|
serine peptidase inhibitor, Kazal type 13 (putative) |
| chr1_+_281863918 | 3.35 |
ENSRNOT00000030108
|
Snu13
|
SNU13 homolog, small nuclear ribonucleoprotein (U4/U6.U5) |
| chr17_-_76281660 | 3.28 |
ENSRNOT00000058070
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr2_-_27343998 | 3.24 |
ENSRNOT00000091418
|
Polk
|
DNA polymerase kappa |
| chr8_-_21224737 | 3.18 |
ENSRNOT00000067321
|
Or7e24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr3_-_48604097 | 3.06 |
ENSRNOT00000009620
|
Ifih1
|
interferon induced with helicase C domain 1 |
| chr3_-_102996314 | 2.99 |
ENSRNOT00000046265
|
Olr774
|
olfactory receptor 774 |
| chr3_-_75576520 | 2.99 |
ENSRNOT00000083330
|
Olr567
|
olfactory receptor 567 |
| chr10_+_82032656 | 2.93 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr2_+_100677411 | 2.88 |
ENSRNOT00000067556
|
Ythdf3
|
YTH N(6)-methyladenosine RNA binding protein 3 |
| chr4_+_146598413 | 2.85 |
ENSRNOT00000067532
|
Atg7
|
autophagy related 7 |
| chr4_+_52235811 | 2.76 |
ENSRNOT00000060478
|
Hyal5
|
hyaluronoglucosaminidase 5 |
| chr1_+_227892956 | 2.66 |
ENSRNOT00000028483
|
AABR07006278.1
|
|
| chr10_+_10457320 | 2.60 |
ENSRNOT00000003832
|
Rbm25l1
|
RNA binding motif protein 25-like 1 |
| chr20_-_34679779 | 2.53 |
ENSRNOT00000078420
|
Cep85l
|
centrosomal protein 85-like |
| chr9_-_121972055 | 2.50 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr3_-_78271875 | 2.49 |
ENSRNOT00000008475
|
Olr701
|
olfactory receptor 701 |
| chr10_-_74388787 | 2.40 |
ENSRNOT00000008107
|
Smg8
|
SMG8 nonsense mediated mRNA decay factor |
| chr7_-_8810649 | 2.31 |
ENSRNOT00000050023
|
Olr937
|
olfactory receptor 937 |
| chr3_-_111587151 | 2.23 |
ENSRNOT00000007299
|
Rpap1
|
RNA polymerase II associated protein 1 |
| chr10_-_58771908 | 2.19 |
ENSRNOT00000018808
|
RGD1304728
|
similar to 4933427D14Rik protein |
| chr1_-_37957400 | 2.17 |
ENSRNOT00000085560
ENSRNOT00000079626 ENSRNOT00000071889 |
LOC102549842
|
zinc finger protein 91-like |
| chr7_-_145062956 | 2.16 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr1_-_167992529 | 2.13 |
ENSRNOT00000067344
|
LOC684170
|
similar to olfactory receptor 566 |
| chr8_-_77398156 | 2.13 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr1_-_22452301 | 2.06 |
ENSRNOT00000043436
|
Taar7h
|
trace amine-associated receptor 7h |
| chr1_+_168005316 | 2.03 |
ENSRNOT00000020678
|
Olr51
|
olfactory receptor 51 |
| chr1_+_222844144 | 2.03 |
ENSRNOT00000032879
|
Pla2g16
|
phospholipase A2, group XVI |
| chr14_-_66771750 | 2.02 |
ENSRNOT00000005587
|
Pacrgl
|
PARK2 co-regulated-like |
| chr7_+_5324469 | 2.00 |
ENSRNOT00000071164
|
Olr901
|
olfactory receptor 901 |
| chrX_-_143558521 | 1.99 |
ENSRNOT00000056598
|
LOC688842
|
hypothetical protein LOC688842 |
| chr15_+_27207645 | 1.95 |
ENSRNOT00000011549
|
Olr1614
|
olfactory receptor 1614 |
| chr9_-_117867557 | 1.93 |
ENSRNOT00000056321
|
AABR07068760.1
|
|
| chr3_+_76237607 | 1.93 |
ENSRNOT00000067444
|
Olr606
|
olfactory receptor 606 |
| chr2_-_61698007 | 1.92 |
ENSRNOT00000049559
|
LOC499541
|
LRRGT00045 |
| chr17_+_53965562 | 1.83 |
ENSRNOT00000090387
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr9_+_42945358 | 1.64 |
ENSRNOT00000059806
|
Fer1l5
|
fer-1-like family member 5 |
| chr13_-_83457888 | 1.37 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr10_-_63240332 | 1.30 |
ENSRNOT00000005125
|
Blmh
|
bleomycin hydrolase |
| chr1_-_173456488 | 1.28 |
ENSRNOT00000044753
|
Olr282
|
olfactory receptor 282 |
| chr19_+_32770655 | 1.22 |
ENSRNOT00000071471
|
LOC100360750
|
Sm protein F-like |
| chr3_-_102826379 | 1.19 |
ENSRNOT00000073423
|
Olr769
|
olfactory receptor 769 |
| chr9_-_105282292 | 1.17 |
ENSRNOT00000071358
|
LOC102554121
|
solute carrier organic anion transporter family member 6A1-like |
| chr3_-_73523043 | 1.10 |
ENSRNOT00000012959
|
Olr484
|
olfactory receptor 484 |
| chr12_+_48677905 | 1.08 |
ENSRNOT00000083196
|
1700069L16Rik
|
RIKEN cDNA 1700069L16 gene |
| chr7_+_116671948 | 1.08 |
ENSRNOT00000077773
ENSRNOT00000029711 |
Gli4
|
GLI family zinc finger 4 |
| chr3_-_76289390 | 1.05 |
ENSRNOT00000051968
|
Olr609
|
olfactory receptor 609 |
| chr10_-_12777245 | 1.03 |
ENSRNOT00000041708
|
Olr1380
|
olfactory receptor 1380 |
| chr3_+_16117228 | 1.00 |
ENSRNOT00000049590
|
Olr404
|
olfactory receptor 404 |
| chr4_+_1566448 | 0.99 |
ENSRNOT00000088227
|
Olr1243
|
olfactory receptor 1243 |
| chrX_-_120521871 | 0.93 |
ENSRNOT00000080863
|
LOC100362376
|
LRRGT00025-like |
| chr5_+_162351001 | 0.92 |
ENSRNOT00000065471
|
LOC691162
|
hypothetical protein LOC691162 |
| chr19_+_22699808 | 0.90 |
ENSRNOT00000023169
|
RGD1308706
|
similar to RIKEN cDNA 4921524J17 |
| chr7_-_101214630 | 0.89 |
ENSRNOT00000042213
|
LOC362921
|
Ac2-224 |
| chr1_-_22404002 | 0.82 |
ENSRNOT00000044098
|
Taar8a
|
trace amine-associated receptor 8a |
| chr13_-_34251650 | 0.80 |
ENSRNOT00000068524
|
Tsn
|
translin |
| chr1_+_42121636 | 0.79 |
ENSRNOT00000025616
|
Myct1
|
myc target 1 |
| chr6_+_29061618 | 0.75 |
ENSRNOT00000092223
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr3_-_61488696 | 0.75 |
ENSRNOT00000083045
|
Lnpk
|
lunapark, ER junction formation factor |
| chr1_+_229722768 | 0.60 |
ENSRNOT00000017126
|
Olr339
|
olfactory receptor 339 |
| chr8_+_118525682 | 0.56 |
ENSRNOT00000028288
|
Elp6
|
elongator acetyltransferase complex subunit 6 |
| chr17_+_39509516 | 0.56 |
ENSRNOT00000022797
|
Prl8a7
|
prolactin family 8, subfamily a, member 7 |
| chr10_-_98544447 | 0.50 |
ENSRNOT00000073149
|
Abca6
|
ATP binding cassette subfamily A member 6 |
| chr1_-_84950210 | 0.47 |
ENSRNOT00000083050
|
AABR07002784.1
|
|
| chr8_+_128577080 | 0.45 |
ENSRNOT00000024172
|
Wdr48
|
WD repeat domain 48 |
| chr8_+_40258985 | 0.44 |
ENSRNOT00000044652
|
Olr1196
|
olfactory receptor 1196 |
| chr16_-_84406538 | 0.39 |
ENSRNOT00000051980
|
LOC500845
|
LRRG00131 |
| chr7_+_6068010 | 0.39 |
ENSRNOT00000044587
|
Olr1007
|
olfactory receptor 1007 |
| chr2_+_149899538 | 0.33 |
ENSRNOT00000065365
|
RGD1560324
|
similar to hypothetical protein C130079G13 |
| chr9_-_8899144 | 0.28 |
ENSRNOT00000051043
|
AABR07066440.1
|
|
| chr2_-_3124543 | 0.27 |
ENSRNOT00000036547
|
Fam81b
|
family with sequence similarity 81, member B |
| chr8_+_18926418 | 0.22 |
ENSRNOT00000044686
|
Olr1128
|
olfactory receptor 1128 |
| chr13_-_34251942 | 0.15 |
ENSRNOT00000044095
|
Tsn
|
translin |
| chr7_+_73273985 | 0.14 |
ENSRNOT00000077730
|
Pop1
|
POP1 homolog, ribonuclease P/MRP subunit |
| chr3_-_103090390 | 0.12 |
ENSRNOT00000045645
|
Olr777
|
olfactory receptor 777 |
| chr3_+_76052230 | 0.07 |
ENSRNOT00000039963
|
LOC100911380
|
olfactory receptor 5W2-like |
| chr3_+_31802999 | 0.02 |
ENSRNOT00000041305
|
AABR07051996.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox21
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.0 | 55.9 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 8.5 | 25.4 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 5.7 | 17.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 3.6 | 18.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 3.0 | 18.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 2.6 | 7.7 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 2.4 | 9.7 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 2.4 | 32.9 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 2.3 | 6.9 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 2.2 | 13.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 2.2 | 6.5 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 2.2 | 8.6 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 2.1 | 8.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 1.8 | 7.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.7 | 7.0 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 1.7 | 5.0 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 1.6 | 6.6 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 1.6 | 7.8 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 1.4 | 4.3 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 1.3 | 5.0 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 1.2 | 29.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 1.2 | 5.8 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 1.2 | 11.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.1 | 11.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 1.0 | 19.9 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 1.0 | 32.5 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.9 | 2.8 | GO:0080144 | amino acid homeostasis(GO:0080144) |
| 0.9 | 23.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.8 | 9.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.8 | 5.3 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.7 | 3.7 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.7 | 3.7 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.7 | 2.1 | GO:0010034 | response to acetate(GO:0010034) |
| 0.6 | 3.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.6 | 1.8 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.5 | 3.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.5 | 4.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.5 | 3.4 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.5 | 3.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.5 | 3.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.4 | 4.7 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.4 | 7.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.4 | 2.8 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.4 | 3.1 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.3 | 5.6 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 13.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.3 | 9.0 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.3 | 6.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.3 | 1.3 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.2 | 18.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.2 | 11.7 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.2 | 55.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.2 | 5.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 2.0 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.2 | 5.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.4 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.1 | 1.8 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 7.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.8 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 2.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 43.9 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.1 | GO:0016078 | tRNA 5'-leader removal(GO:0001682) tRNA catabolic process(GO:0016078) |
| 0.0 | 3.8 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 1.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 3.9 | GO:0008380 | RNA splicing(GO:0008380) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 32.5 | GO:0014731 | spectrin(GO:0008091) spectrin-associated cytoskeleton(GO:0014731) cuticular plate(GO:0032437) |
| 2.8 | 8.3 | GO:0043511 | inhibin complex(GO:0043511) |
| 2.3 | 11.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 2.0 | 34.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 1.3 | 16.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 1.2 | 17.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.8 | 9.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.7 | 3.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.7 | 13.7 | GO:0001741 | XY body(GO:0001741) |
| 0.6 | 4.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 3.3 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.4 | 7.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.4 | 5.0 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.3 | 6.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 17.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 7.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 11.3 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.2 | 13.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 7.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 90.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.2 | 3.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 3.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 9.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 2.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 23.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 76.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 5.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 49.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 4.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 10.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 23.5 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 2.0 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 7.6 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 2.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 2.5 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 3.2 | GO:0031252 | cell leading edge(GO:0031252) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 23.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 3.6 | 14.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 3.0 | 18.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 2.6 | 7.7 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 2.5 | 5.0 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 2.3 | 6.9 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 2.2 | 6.5 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 2.1 | 8.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.9 | 9.7 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 1.5 | 35.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 1.2 | 3.7 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 1.2 | 5.8 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 1.1 | 9.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 1.1 | 3.3 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 1.1 | 17.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.9 | 5.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.8 | 14.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.7 | 11.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.5 | 6.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.5 | 62.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 2.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 1.8 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.4 | 2.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 2.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.3 | 7.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 2.8 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.2 | 36.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 17.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 25.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 6.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 3.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 21.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.2 | 6.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 5.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 2.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 18.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 7.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 4.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 31.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 3.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 8.7 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 9.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 27.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 46.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 3.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 3.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 14.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 6.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 4.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.5 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.5 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 73.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.9 | 26.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.4 | 11.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.4 | 3.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.4 | 84.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 13.7 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 8.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 12.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 7.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 7.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 4.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 6.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 5.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 45.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 1.2 | 23.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 1.2 | 33.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.9 | 30.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 5.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.8 | 8.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.7 | 30.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.6 | 14.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.6 | 7.0 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.5 | 3.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.5 | 10.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.5 | 18.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.4 | 5.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 9.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.3 | 6.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.3 | 7.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 9.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 4.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 3.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 5.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 9.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 3.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 3.7 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |