Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Sox18_Sox12
Z-value: 1.00
Transcription factors associated with Sox18_Sox12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
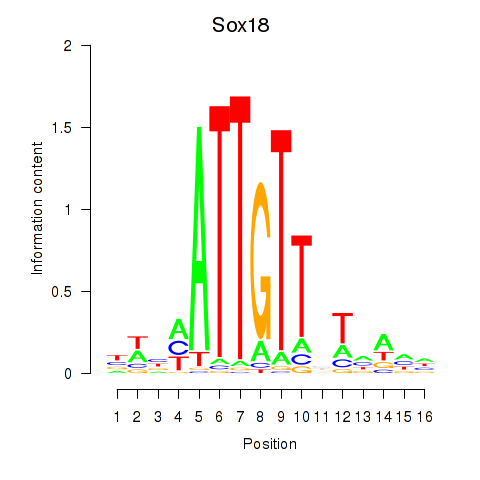
Sox18
|
ENSRNOG00000016248 | SRY box 18 |
|
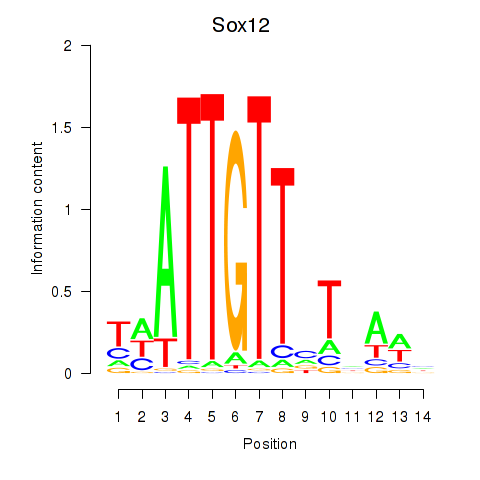
Sox12
|
ENSRNOG00000007514 | SRY box 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox18 | rn6_v1_chr3_-_177179127_177179127 | 0.44 | 9.5e-17 | Click! |
| Sox12 | rn6_v1_chr3_-_147865393_147865393 | -0.04 | 4.3e-01 | Click! |
Activity profile of Sox18_Sox12 motif
Sorted Z-values of Sox18_Sox12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_12915734 | 37.98 |
ENSRNOT00000089066
|
Txlnb
|
taxilin beta |
| chrX_+_159158194 | 37.41 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr3_-_37803112 | 30.94 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr6_-_23316962 | 27.85 |
ENSRNOT00000065421
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr9_-_97290639 | 25.80 |
ENSRNOT00000026491
ENSRNOT00000056724 |
Iqca1
|
IQ motif containing with AAA domain 1 |
| chr13_-_93307199 | 24.62 |
ENSRNOT00000065041
|
Rgs7
|
regulator of G-protein signaling 7 |
| chr2_+_22950018 | 23.73 |
ENSRNOT00000071804
|
Homer1
|
homer scaffolding protein 1 |
| chr2_-_191294374 | 23.02 |
ENSRNOT00000067469
|
RGD1562234
|
similar to S100 calcium-binding protein, ventral prostate |
| chr3_-_52849907 | 22.63 |
ENSRNOT00000041096
|
Scn7a
|
sodium voltage-gated channel alpha subunit 7 |
| chr2_+_248398917 | 19.22 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr4_-_183544850 | 17.98 |
ENSRNOT00000071407
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr13_+_57243877 | 17.88 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr11_+_80736576 | 15.67 |
ENSRNOT00000047678
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr6_+_73553210 | 15.23 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr2_+_54466280 | 14.53 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr3_+_139894331 | 14.33 |
ENSRNOT00000064695
|
Rin2
|
Ras and Rab interactor 2 |
| chr13_+_87986240 | 14.02 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr3_+_92640752 | 13.78 |
ENSRNOT00000007604
|
Slc1a2
|
solute carrier family 1 member 2 |
| chr10_-_98544447 | 13.74 |
ENSRNOT00000073149
|
Abca6
|
ATP binding cassette subfamily A member 6 |
| chr16_+_23553647 | 13.70 |
ENSRNOT00000041994
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr4_+_21462779 | 13.29 |
ENSRNOT00000089747
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr7_-_3707226 | 13.24 |
ENSRNOT00000064380
|
Olr879
|
olfactory receptor 879 |
| chr6_+_55371738 | 13.22 |
ENSRNOT00000006652
|
Agr3
|
anterior gradient 3, protein disulphide isomerase family member |
| chr5_+_90338795 | 12.95 |
ENSRNOT00000077864
ENSRNOT00000058882 |
LOC298139
|
similar to RIKEN cDNA 2310003M01 |
| chr18_+_30010918 | 12.59 |
ENSRNOT00000084132
|
Pcdha4
|
protocadherin alpha 4 |
| chr15_-_50425900 | 12.50 |
ENSRNOT00000058715
|
Adam28
|
ADAM metallopeptidase domain 28 |
| chr4_-_166803127 | 12.08 |
ENSRNOT00000067909
|
RGD1306750
|
LOC362451 |
| chr18_+_30550877 | 12.06 |
ENSRNOT00000027164
|
LOC108348201
|
protocadherin beta-7-like |
| chr15_+_10120206 | 11.96 |
ENSRNOT00000033048
|
Rarb
|
retinoic acid receptor, beta |
| chr1_+_114679537 | 11.93 |
ENSRNOT00000019498
|
Oca2
|
OCA2 melanosomal transmembrane protein |
| chr3_+_128155069 | 11.66 |
ENSRNOT00000051184
ENSRNOT00000006389 |
Plcb1
|
phospholipase C beta 1 |
| chr4_-_14490446 | 11.45 |
ENSRNOT00000009132
|
Sema3c
|
semaphorin 3C |
| chr2_-_173563273 | 11.33 |
ENSRNOT00000081423
|
Zbbx
|
zinc finger, B-box domain containing |
| chr17_+_33028293 | 11.25 |
ENSRNOT00000072764
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr16_+_29674793 | 11.15 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr1_-_76780230 | 10.80 |
ENSRNOT00000002046
|
LOC100912485
|
alcohol sulfotransferase-like |
| chr2_-_192642548 | 10.47 |
ENSRNOT00000030295
|
Sprr3
|
small proline-rich protein 3 |
| chr8_-_84522588 | 10.45 |
ENSRNOT00000076213
|
Mlip
|
muscular LMNA-interacting protein |
| chr13_-_50761306 | 10.31 |
ENSRNOT00000076610
ENSRNOT00000004241 |
Prelp
|
proline and arginine rich end leucine rich repeat protein |
| chr3_-_21027947 | 10.10 |
ENSRNOT00000051973
|
Olr421
|
olfactory receptor 421 |
| chr18_+_30435119 | 10.05 |
ENSRNOT00000027190
|
Pcdhb8
|
protocadherin beta 8 |
| chr18_+_30496318 | 9.85 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr15_-_52214616 | 9.83 |
ENSRNOT00000015035
|
Sftpc
|
surfactant protein C |
| chrM_+_7758 | 9.82 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr6_-_23291568 | 9.73 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr17_+_9736577 | 9.71 |
ENSRNOT00000066586
|
F12
|
coagulation factor XII |
| chr5_+_15043955 | 9.62 |
ENSRNOT00000047093
|
Rp1
|
retinitis pigmentosa 1 |
| chr4_-_89695928 | 9.57 |
ENSRNOT00000039316
|
Gprin3
|
GPRIN family member 3 |
| chr5_+_146656049 | 9.56 |
ENSRNOT00000036029
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr11_-_782954 | 9.50 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr1_-_128287151 | 9.49 |
ENSRNOT00000084946
ENSRNOT00000089723 |
Mef2a
|
myocyte enhancer factor 2a |
| chr1_+_177093387 | 9.40 |
ENSRNOT00000021858
|
Mical2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr4_+_96562725 | 9.27 |
ENSRNOT00000009094
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr14_-_19191863 | 8.73 |
ENSRNOT00000003921
|
Alb
|
albumin |
| chr4_+_14151343 | 8.64 |
ENSRNOT00000061687
ENSRNOT00000076573 ENSRNOT00000077219 ENSRNOT00000008319 |
Cd36
|
CD36 molecule |
| chr18_+_30562178 | 8.60 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr16_+_31734944 | 8.53 |
ENSRNOT00000059673
|
Palld
|
palladin, cytoskeletal associated protein |
| chr2_-_182035032 | 8.52 |
ENSRNOT00000009813
|
Fgb
|
fibrinogen beta chain |
| chr4_+_155321553 | 8.45 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr6_+_50725085 | 8.29 |
ENSRNOT00000009473
|
Slc26a3
|
solute carrier family 26 member 3 |
| chr16_-_19791832 | 8.24 |
ENSRNOT00000040393
|
Ushbp1
|
USH1 protein network component harmonin binding protein 1 |
| chr1_-_167911961 | 8.21 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr2_-_117454769 | 8.19 |
ENSRNOT00000068381
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr3_-_94182714 | 7.89 |
ENSRNOT00000015073
|
LOC100362814
|
hypothetical protein LOC100362814 |
| chr2_+_78245459 | 7.86 |
ENSRNOT00000089551
|
LOC103689968
|
protein FAM134B |
| chr17_+_9736786 | 7.80 |
ENSRNOT00000081920
|
F12
|
coagulation factor XII |
| chr4_+_71621729 | 7.72 |
ENSRNOT00000022275
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr18_+_52550739 | 7.72 |
ENSRNOT00000037529
|
Ctxn3
|
cortexin 3 |
| chr10_+_82775691 | 7.67 |
ENSRNOT00000030737
|
Hils1
|
histone linker H1 domain, spermatid-specific 1 |
| chrX_+_118197217 | 7.59 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr7_-_57679795 | 7.54 |
ENSRNOT00000007461
|
Trhde
|
thyrotropin-releasing hormone degrading enzyme |
| chr3_+_86621205 | 7.50 |
ENSRNOT00000044595
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr12_-_5685448 | 7.31 |
ENSRNOT00000077167
|
Fry
|
FRY microtubule binding protein |
| chr6_+_107603580 | 7.16 |
ENSRNOT00000036430
|
Dnal1
|
dynein, axonemal, light chain 1 |
| chr3_-_72219246 | 7.12 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr10_-_54512169 | 7.07 |
ENSRNOT00000005066
|
Cfap52
|
cilia and flagella associated protein 52 |
| chr6_+_110624856 | 7.03 |
ENSRNOT00000014017
|
Vash1
|
vasohibin 1 |
| chr12_-_42492328 | 7.01 |
ENSRNOT00000011552
|
Tbx3
|
T-box 3 |
| chrX_-_142248369 | 6.94 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr8_-_87282156 | 6.94 |
ENSRNOT00000087874
|
Filip1
|
filamin A interacting protein 1 |
| chr11_+_61083757 | 6.91 |
ENSRNOT00000002790
|
Boc
|
BOC cell adhesion associated, oncogene regulated |
| chr18_-_75207306 | 6.90 |
ENSRNOT00000021717
|
Setbp1
|
SET binding protein 1 |
| chr15_-_39886613 | 6.86 |
ENSRNOT00000089963
|
Cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr3_+_110669312 | 6.80 |
ENSRNOT00000013829
|
Ivd
|
isovaleryl-CoA dehydrogenase |
| chr9_+_118842787 | 6.77 |
ENSRNOT00000090512
|
Dlgap1
|
DLG associated protein 1 |
| chr5_+_64476317 | 6.77 |
ENSRNOT00000017217
|
LOC108348074
|
collagen alpha-1(XV) chain-like |
| chr18_+_30509393 | 6.72 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr8_-_73194837 | 6.62 |
ENSRNOT00000024885
ENSRNOT00000081450 ENSRNOT00000064613 |
Tln2
|
talin 2 |
| chr12_-_42492526 | 6.60 |
ENSRNOT00000084018
|
Tbx3
|
T-box 3 |
| chr16_+_54153054 | 6.53 |
ENSRNOT00000090644
ENSRNOT00000014248 |
Fgl1
|
fibrinogen-like 1 |
| chr17_+_60059949 | 6.50 |
ENSRNOT00000025458
|
Mpp7
|
membrane palmitoylated protein 7 |
| chr1_-_258766881 | 6.46 |
ENSRNOT00000015801
|
Cyp2c12
|
cytochrome P450, family 2, subfamily c, polypeptide 12 |
| chr17_-_32076181 | 6.44 |
ENSRNOT00000074842
|
LOC100911107
|
leukocyte elastase inhibitor A-like |
| chr15_-_108120279 | 6.27 |
ENSRNOT00000090572
|
Dock9
|
dedicator of cytokinesis 9 |
| chr2_-_18531210 | 6.24 |
ENSRNOT00000088313
|
Vcan
|
versican |
| chr11_+_42259761 | 6.20 |
ENSRNOT00000047310
|
Epha6
|
Eph receptor A6 |
| chr3_-_57913999 | 6.18 |
ENSRNOT00000090426
|
Slc25a12
|
solute carrier family 25 member 12 |
| chr18_+_17091310 | 6.16 |
ENSRNOT00000093285
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr2_-_1511591 | 6.07 |
ENSRNOT00000087906
ENSRNOT00000014709 |
Cast
|
calpastatin |
| chr14_+_33354807 | 5.98 |
ENSRNOT00000049774
|
Hopx
|
HOP homeobox |
| chr16_+_23317953 | 5.82 |
ENSRNOT00000075287
|
AABR07024972.1
|
|
| chr12_+_10636275 | 5.78 |
ENSRNOT00000001285
|
Cyp3a18
|
cytochrome P450, family 3, subfamily a, polypeptide 18 |
| chrM_+_10160 | 5.77 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chrX_+_6273733 | 5.75 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr9_+_81631409 | 5.70 |
ENSRNOT00000089580
|
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr16_+_54164431 | 5.58 |
ENSRNOT00000090763
|
Fgl1
|
fibrinogen-like 1 |
| chr18_+_30381322 | 5.57 |
ENSRNOT00000027216
|
Pcdhb3
|
protocadherin beta 3 |
| chr1_-_275882444 | 5.55 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr11_-_31772984 | 5.53 |
ENSRNOT00000002771
|
Dnajc28
|
DnaJ heat shock protein family (Hsp40) member C28 |
| chr9_+_52023295 | 5.52 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr9_+_74124016 | 5.49 |
ENSRNOT00000019023
|
Cps1
|
carbamoyl-phosphate synthase 1 |
| chr1_-_76517134 | 5.41 |
ENSRNOT00000064593
ENSRNOT00000085775 |
LOC100912485
|
alcohol sulfotransferase-like |
| chr5_+_116420690 | 5.40 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr2_-_158156444 | 5.40 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr13_+_6679368 | 5.37 |
ENSRNOT00000067198
ENSRNOT00000092965 |
Cntnap5c
|
contactin associated protein-like 5C |
| chr11_+_7265828 | 5.33 |
ENSRNOT00000084765
|
Gbe1
|
1,4-alpha-glucan branching enzyme 1 |
| chr18_+_29993361 | 5.32 |
ENSRNOT00000075810
|
Pcdha4
|
protocadherin alpha 4 |
| chr9_-_112027155 | 5.32 |
ENSRNOT00000021258
ENSRNOT00000080962 |
Pja2
|
praja ring finger ubiquitin ligase 2 |
| chr13_+_24823488 | 5.32 |
ENSRNOT00000019907
|
Cdh20
|
cadherin 20 |
| chr14_+_115275894 | 5.16 |
ENSRNOT00000033437
|
Gpr75
|
G protein-coupled receptor 75 |
| chr3_+_51687809 | 5.12 |
ENSRNOT00000087242
|
Scn2a
|
sodium voltage-gated channel alpha subunit 2 |
| chr3_-_160038078 | 5.11 |
ENSRNOT00000013445
|
Serinc3
|
serine incorporator 3 |
| chr11_-_4332255 | 5.05 |
ENSRNOT00000087133
|
Cadm2
|
cell adhesion molecule 2 |
| chr6_-_54020406 | 5.01 |
ENSRNOT00000086038
|
Hdac9
|
histone deacetylase 9 |
| chr5_-_168734296 | 4.95 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr5_+_117698764 | 4.90 |
ENSRNOT00000011486
|
Angptl3
|
angiopoietin-like 3 |
| chr1_+_21613148 | 4.89 |
ENSRNOT00000018695
|
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chrX_+_107496072 | 4.89 |
ENSRNOT00000003283
|
Plp1
|
proteolipid protein 1 |
| chrX_+_10042893 | 4.83 |
ENSRNOT00000084218
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr13_-_88061108 | 4.78 |
ENSRNOT00000003774
|
Rgs4
|
regulator of G-protein signaling 4 |
| chr16_+_25773602 | 4.78 |
ENSRNOT00000047750
|
Gapdh-ps2
|
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 2 |
| chrM_+_3904 | 4.76 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr15_+_18451144 | 4.73 |
ENSRNOT00000010260
|
Acox2
|
acyl-CoA oxidase 2 |
| chr8_-_17525906 | 4.67 |
ENSRNOT00000007855
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr8_+_100260049 | 4.67 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr5_+_104394119 | 4.66 |
ENSRNOT00000093557
ENSRNOT00000093462 |
Adamtsl1
|
ADAMTS-like 1 |
| chr14_+_114152472 | 4.58 |
ENSRNOT00000042965
|
Rtn4
|
reticulon 4 |
| chr2_-_216443518 | 4.51 |
ENSRNOT00000022496
|
Amy1a
|
amylase, alpha 1A |
| chr3_+_69549673 | 4.49 |
ENSRNOT00000043974
|
Zfp804a
|
zinc finger protein 804A |
| chr2_-_183213228 | 4.47 |
ENSRNOT00000067618
|
Trim2
|
tripartite motif-containing 2 |
| chr5_-_50193571 | 4.46 |
ENSRNOT00000051243
|
Cfap206
|
cilia and flagella associated protein 206 |
| chr3_+_171832500 | 4.40 |
ENSRNOT00000007554
|
Vapb
|
VAMP associated protein B and C |
| chr18_+_30474947 | 4.38 |
ENSRNOT00000027188
|
Pcdhb9
|
protocadherin beta 9 |
| chrM_+_7919 | 4.38 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr18_-_11906827 | 4.35 |
ENSRNOT00000078207
|
Dsc1
|
desmocollin 1 |
| chr5_-_166726794 | 4.25 |
ENSRNOT00000022799
|
Slc25a33
|
solute carrier family 25 member 33 |
| chr20_+_3344870 | 4.24 |
ENSRNOT00000001058
ENSRNOT00000087613 ENSRNOT00000089978 |
Mrps18b
|
mitochondrial ribosomal protein S18B |
| chr1_-_213635546 | 4.18 |
ENSRNOT00000018861
|
Sirt3
|
sirtuin 3 |
| chr2_-_113345577 | 4.16 |
ENSRNOT00000034096
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr18_-_62476700 | 4.16 |
ENSRNOT00000048429
|
Cycs
|
cytochrome c, somatic |
| chr14_-_28967980 | 4.13 |
ENSRNOT00000048175
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr2_-_28799266 | 4.13 |
ENSRNOT00000089293
|
Tmem171
|
transmembrane protein 171 |
| chr4_+_31534225 | 4.12 |
ENSRNOT00000089292
ENSRNOT00000016423 |
NEWGENE_621351
|
collagen, type I, alpha 2 |
| chr9_+_81816872 | 4.10 |
ENSRNOT00000041407
|
Plcd4
|
phospholipase C, delta 4 |
| chr2_+_193286721 | 4.09 |
ENSRNOT00000074852
|
AABR07012323.1
|
|
| chr20_+_41266566 | 4.09 |
ENSRNOT00000000653
|
Frk
|
fyn-related Src family tyrosine kinase |
| chrX_+_84064427 | 4.07 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr3_+_60024013 | 4.04 |
ENSRNOT00000025255
|
Scrn3
|
secernin 3 |
| chr4_-_108008484 | 4.03 |
ENSRNOT00000007971
|
Ctnna2
|
catenin alpha 2 |
| chr3_+_104042168 | 3.98 |
ENSRNOT00000065752
|
Aven
|
apoptosis and caspase activation inhibitor |
| chr11_+_65960277 | 3.96 |
ENSRNOT00000003674
|
Ndufb4
|
NADH:ubiquinone oxidoreductase subunit B4 |
| chr3_-_77800137 | 3.96 |
ENSRNOT00000071319
|
AABR07052831.1
|
|
| chr1_+_213577122 | 3.95 |
ENSRNOT00000071925
|
RGD1309350
|
similar to transthyretin (4L369) |
| chr4_-_11610518 | 3.94 |
ENSRNOT00000066643
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr14_-_114484127 | 3.92 |
ENSRNOT00000056706
|
Eml6
|
echinoderm microtubule associated protein like 6 |
| chr2_-_35503638 | 3.89 |
ENSRNOT00000007560
|
LOC100910467
|
olfactory receptor 144-like |
| chrX_+_121612952 | 3.86 |
ENSRNOT00000022122
|
AABR07041179.1
|
|
| chr19_+_487723 | 3.85 |
ENSRNOT00000061734
|
Ces2j
|
carboxylesterase 2J |
| chr5_-_127878792 | 3.82 |
ENSRNOT00000014436
|
Zyg11b
|
zyg-11 family member B, cell cycle regulator |
| chr5_-_136965191 | 3.79 |
ENSRNOT00000056842
|
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr18_+_30387937 | 3.73 |
ENSRNOT00000027210
|
Pcdhb4
|
protocadherin beta 4 |
| chr1_-_171009670 | 3.70 |
ENSRNOT00000026487
|
Olr222
|
olfactory receptor 222 |
| chrX_+_110789269 | 3.68 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr8_-_38549268 | 3.59 |
ENSRNOT00000088001
|
LOC100912026
|
urinary protein 3-like |
| chr8_+_57983556 | 3.51 |
ENSRNOT00000009562
|
RGD1311251
|
similar to RIKEN cDNA 4930550C14 |
| chr3_-_77696294 | 3.46 |
ENSRNOT00000050849
|
Olr670
|
olfactory receptor 670 |
| chr13_-_68360664 | 3.46 |
ENSRNOT00000030971
|
Hmcn1
|
hemicentin 1 |
| chr9_+_24066303 | 3.46 |
ENSRNOT00000018163
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr13_-_68253251 | 3.44 |
ENSRNOT00000076776
|
Hmcn1
|
hemicentin 1 |
| chr5_+_10178302 | 3.40 |
ENSRNOT00000009679
|
Sntg1
|
syntrophin, gamma 1 |
| chr18_+_29951094 | 3.33 |
ENSRNOT00000027402
|
LOC102553180
|
protocadherin alpha-1-like |
| chr2_-_186245771 | 3.32 |
ENSRNOT00000066004
ENSRNOT00000079954 |
Dclk2
|
doublecortin-like kinase 2 |
| chr20_-_29579578 | 3.31 |
ENSRNOT00000000700
|
AC096404.1
|
|
| chr11_+_57265732 | 3.31 |
ENSRNOT00000093174
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr7_-_15163866 | 3.30 |
ENSRNOT00000042638
|
Zfp870
|
zinc finger protein 870 |
| chr9_+_47134034 | 3.26 |
ENSRNOT00000020108
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr3_+_77261334 | 3.21 |
ENSRNOT00000049527
|
LOC103690355
|
olfactory receptor 4C11-like |
| chr2_+_85305225 | 3.19 |
ENSRNOT00000015904
|
Tas2r119
|
taste receptor, type 2, member 119 |
| chr5_+_172825072 | 3.08 |
ENSRNOT00000068525
|
Cfap74
|
cilia and flagella associated protein 74 |
| chr2_-_44981458 | 3.07 |
ENSRNOT00000014134
|
Gzma
|
granzyme A |
| chr8_-_18508971 | 3.05 |
ENSRNOT00000061339
|
Muc16
|
mucin 16, cell surface associated |
| chr7_-_114573900 | 3.05 |
ENSRNOT00000011219
|
Ptk2
|
protein tyrosine kinase 2 |
| chr3_+_103597194 | 3.04 |
ENSRNOT00000073004
|
LOC502660
|
similar to olfactory receptor 1318 |
| chr1_-_150210403 | 3.03 |
ENSRNOT00000081156
|
Olr29
|
olfactory receptor 29 |
| chr1_-_67065797 | 3.01 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr4_+_136676254 | 3.01 |
ENSRNOT00000088070
|
Cntn6
|
contactin 6 |
| chr7_+_6068010 | 3.00 |
ENSRNOT00000044587
|
Olr1007
|
olfactory receptor 1007 |
| chr7_-_59882077 | 2.97 |
ENSRNOT00000068774
|
Myrfl
|
myelin regulatory factor-like |
| chr5_-_75910805 | 2.92 |
ENSRNOT00000019099
|
Olr854
|
olfactory receptor 854 |
| chr3_+_75465399 | 2.89 |
ENSRNOT00000057479
|
Olr562
|
olfactory receptor 562 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox18_Sox12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.5 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 4.8 | 14.5 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 4.5 | 13.6 | GO:0060931 | sinoatrial node cell development(GO:0060931) His-Purkinje system cell differentiation(GO:0060932) |
| 3.9 | 11.7 | GO:2000437 | interleukin-12-mediated signaling pathway(GO:0035722) interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-12(GO:0071349) cellular response to interleukin-15(GO:0071350) regulation of monocyte extravasation(GO:2000437) |
| 3.8 | 11.5 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 3.8 | 15.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 3.2 | 19.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 3.2 | 9.5 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 3.2 | 9.5 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 3.1 | 9.4 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 2.8 | 13.8 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 2.7 | 8.2 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 2.5 | 7.6 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 2.3 | 6.8 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 2.2 | 37.0 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 2.1 | 6.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.0 | 9.8 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 1.8 | 5.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.8 | 7.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 1.8 | 7.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 1.7 | 8.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.7 | 10.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 1.6 | 9.8 | GO:0070541 | response to platinum ion(GO:0070541) |
| 1.6 | 4.9 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 1.6 | 9.8 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 1.6 | 6.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.6 | 4.7 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 1.6 | 7.9 | GO:0061709 | reticulophagy(GO:0061709) |
| 1.5 | 12.0 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 1.4 | 15.7 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 1.3 | 5.1 | GO:0009597 | detection of virus(GO:0009597) |
| 1.3 | 2.5 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 1.2 | 9.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.1 | 5.7 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 1.1 | 5.5 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 1.1 | 7.6 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 1.1 | 3.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 1.0 | 4.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 1.0 | 10.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 1.0 | 6.9 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.0 | 4.9 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 1.0 | 6.8 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.9 | 8.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.9 | 2.6 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.9 | 9.6 | GO:0046548 | photoreceptor cell outer segment organization(GO:0035845) retinal rod cell development(GO:0046548) |
| 0.8 | 6.7 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.8 | 37.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.8 | 3.0 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.8 | 12.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.7 | 4.4 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.7 | 5.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.7 | 6.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.7 | 6.0 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.7 | 4.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.6 | 96.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.6 | 1.3 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.6 | 1.9 | GO:2000360 | regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.6 | 5.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.6 | 1.2 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.6 | 6.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.6 | 13.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.6 | 0.6 | GO:1904404 | response to formaldehyde(GO:1904404) |
| 0.5 | 4.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.5 | 2.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.5 | 3.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.5 | 9.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.5 | 1.5 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.5 | 14.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.5 | 8.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.5 | 1.8 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.5 | 10.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 4.0 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.4 | 23.5 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.4 | 0.9 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.4 | 3.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.4 | 2.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.4 | 4.2 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.4 | 7.8 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.4 | 6.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 4.9 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.4 | 4.7 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.4 | 17.9 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.4 | 4.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.4 | 10.4 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.4 | 23.8 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.4 | 5.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.3 | 8.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.3 | 3.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 10.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 1.5 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.3 | 6.9 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.3 | 6.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 7.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.3 | 2.0 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 5.5 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.2 | 1.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 7.7 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 6.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 0.9 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 | 1.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 2.3 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.2 | 8.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.2 | 0.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.2 | 1.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.2 | 5.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.2 | 7.1 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.2 | 1.9 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 1.9 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 7.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 2.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 1.7 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.2 | 3.8 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 2.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.2 | 2.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 0.7 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 0.5 | GO:1990168 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 1.9 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.2 | 2.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 1.0 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 0.3 | GO:0032752 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.2 | 0.9 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 4.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 6.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 2.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 3.9 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 6.2 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 0.3 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 1.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 2.0 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 156.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.9 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) tetrahydrobiopterin biosynthetic process(GO:0006729) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.1 | 1.9 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.9 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 1.9 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 2.3 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 2.1 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.1 | 18.7 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 2.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 1.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 2.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.1 | 0.5 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 4.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 4.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.5 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.1 | 5.0 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 1.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 2.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.2 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.1 | 2.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 3.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 4.9 | GO:0007569 | cell aging(GO:0007569) |
| 0.1 | 2.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 2.6 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.6 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.1 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.5 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 2.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.3 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.0 | 0.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 1.2 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.4 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.9 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 1.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 1.7 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.0 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.0 | 2.3 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.9 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.9 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 1.6 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.0 | 2.1 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 4.8 | 19.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 2.1 | 14.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.6 | 6.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 1.4 | 27.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.3 | 11.9 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 1.2 | 24.6 | GO:0044292 | dendrite terminus(GO:0044292) |
| 1.1 | 8.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.9 | 12.8 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.8 | 9.8 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.8 | 9.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.8 | 8.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.7 | 4.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.7 | 6.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.6 | 13.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.5 | 23.7 | GO:0043034 | costamere(GO:0043034) |
| 0.5 | 18.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.5 | 15.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.5 | 5.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 3.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 6.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.4 | 10.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 8.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.4 | 5.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.3 | 2.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.3 | 2.7 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.3 | 2.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 10.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.3 | 4.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.3 | 16.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 7.1 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 0.9 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.3 | 4.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 27.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.3 | 1.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 6.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 0.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 3.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 4.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 3.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 8.5 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.2 | 41.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 17.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 5.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 2.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 4.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 3.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 5.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 6.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 3.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 2.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 5.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 9.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 8.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 6.3 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 12.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 1.0 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 2.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 3.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 25.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 64.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 9.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 4.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 6.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 4.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 189.7 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 2.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 41.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 2.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 12.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 10.2 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 13.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.9 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 2.7 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 12.8 | GO:0005739 | mitochondrion(GO:0005739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 23.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 4.4 | 13.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 4.1 | 16.6 | GO:0019002 | GMP binding(GO:0019002) |
| 2.8 | 13.8 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 2.5 | 7.6 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 2.0 | 9.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 1.9 | 24.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.8 | 5.3 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 1.7 | 15.2 | GO:0043495 | protein anchor(GO:0043495) |
| 1.6 | 4.9 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 1.5 | 4.5 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.2 | 6.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.1 | 5.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 1.1 | 4.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 1.1 | 3.3 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 1.0 | 12.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 1.0 | 12.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.0 | 3.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.9 | 9.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.9 | 3.8 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.9 | 4.7 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.9 | 27.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.9 | 5.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.9 | 2.7 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.7 | 15.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.7 | 9.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.7 | 16.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.6 | 2.5 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.6 | 6.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.6 | 6.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.6 | 17.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.6 | 2.5 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.6 | 16.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.6 | 3.9 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.6 | 8.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.5 | 4.9 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.5 | 13.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.5 | 6.8 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.5 | 5.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.5 | 1.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.5 | 1.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.5 | 5.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.5 | 11.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.5 | 0.5 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.5 | 8.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 2.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 6.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 7.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.4 | 11.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.4 | 4.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 1.2 | GO:0052870 | leukotriene-B4 20-monooxygenase activity(GO:0050051) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.4 | 2.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.4 | 9.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.4 | 1.9 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.4 | 15.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.4 | 5.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.4 | 2.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.4 | 1.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.4 | 1.1 | GO:0004655 | porphobilinogen synthase activity(GO:0004655) |
| 0.3 | 3.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.3 | 2.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 7.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 2.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 7.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 2.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 0.9 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.3 | 15.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 2.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.3 | 13.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.3 | 36.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.3 | 0.9 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.3 | 4.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.3 | 43.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.3 | 1.7 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.3 | 5.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.3 | 0.8 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.3 | 0.8 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 0.3 | 1.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 9.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 2.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 4.2 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.2 | 1.3 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.2 | 2.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 4.2 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.2 | 2.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 20.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.2 | 1.6 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 3.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 2.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 106.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.2 | 7.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 3.7 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.2 | 12.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.2 | 0.7 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.2 | 6.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 0.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 5.0 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 2.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 164.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.5 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 6.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 3.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 4.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 10.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 2.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 2.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.6 | GO:0072349 | cofactor transporter activity(GO:0051184) modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 12.6 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 8.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 6.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 2.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.3 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 3.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 6.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 6.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 6.1 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 6.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 5.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 3.0 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.9 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 3.3 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 8.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 2.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 9.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.4 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 7.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 2.0 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 1.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.4 | 15.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.4 | 11.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.4 | 6.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.4 | 15.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 8.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.3 | 12.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.3 | 6.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 6.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 35.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 6.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 9.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 6.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 4.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 13.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 8.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 5.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.2 | 3.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 26.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 6.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 33.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 6.6 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 1.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 2.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 5.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 14.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 16.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.2 | 11.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.9 | 8.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.9 | 18.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.8 | 30.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.8 | 27.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 4.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.7 | 10.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.7 | 13.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.7 | 11.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.6 | 14.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.6 | 13.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.5 | 6.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 16.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.5 | 6.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 6.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 4.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.3 | 6.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 17.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 13.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 8.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 10.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 3.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 6.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 5.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 2.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 1.3 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.2 | 4.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 9.8 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 8.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 4.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 4.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 0.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 2.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 10.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 5.3 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 3.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 0.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 0.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.6 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 6.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 6.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 4.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 2.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 3.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |