Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
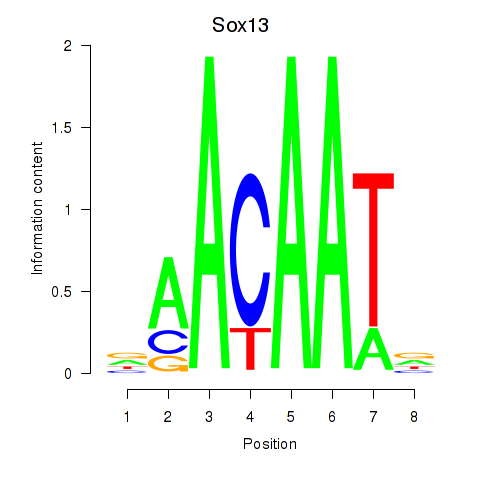
Results for Sox13
Z-value: 0.19
Transcription factors associated with Sox13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox13
|
ENSRNOG00000028353 | SRY box 13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox13 | rn6_v1_chr13_+_50434886_50434886 | -0.04 | 4.7e-01 | Click! |
Activity profile of Sox13 motif
Sorted Z-values of Sox13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_64088900 | 10.12 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr11_+_86520992 | 2.59 |
ENSRNOT00000040954
|
Gp1bb
|
glycoprotein Ib platelet beta subunit |
| chr18_+_30562178 | 2.20 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr4_+_165732643 | 2.05 |
ENSRNOT00000034403
|
LOC690326
|
hypothetical protein LOC690326 |
| chr9_-_112027155 | 1.68 |
ENSRNOT00000021258
ENSRNOT00000080962 |
Pja2
|
praja ring finger ubiquitin ligase 2 |
| chr1_+_234363994 | 1.18 |
ENSRNOT00000018137
|
Rorb
|
RAR-related orphan receptor B |
| chr5_+_103251986 | 0.95 |
ENSRNOT00000008757
|
Cntln
|
centlein |
| chr13_-_78521911 | 0.94 |
ENSRNOT00000087506
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr3_+_102510339 | 0.71 |
ENSRNOT00000074883
|
LOC691368
|
similar to olfactory receptor 1278 |
| chr3_-_134340801 | 0.52 |
ENSRNOT00000085149
|
Sel1l2
|
SEL1L2 ERAD E3 ligase adaptor subunit |
| chr11_-_29638922 | 0.40 |
ENSRNOT00000048888
|
LOC100359671
|
ribosomal protein L29-like |
| chr20_-_43932361 | 0.34 |
ENSRNOT00000091030
|
Rfpl4b
|
ret finger protein-like 4B |
| chr1_+_63263722 | 0.27 |
ENSRNOT00000088203
|
LOC100911184
|
vomeronasal type-1 receptor 4-like |
| chr3_-_5483239 | 0.02 |
ENSRNOT00000071754
|
Surf4
|
surfeit 4 |
| chr8_-_43082948 | 0.02 |
ENSRNOT00000072050
|
LOC100910667
|
olfactory receptor 8G5-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 10.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 1.2 | GO:0046549 | amacrine cell differentiation(GO:0035881) retinal rod cell development(GO:0046548) retinal cone cell development(GO:0046549) |
| 0.1 | 1.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 2.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 2.6 | GO:0007596 | blood coagulation(GO:0007596) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.1 | GO:0032280 | symmetric synapse(GO:0032280) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.2 | 1.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 1.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 2.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |