Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
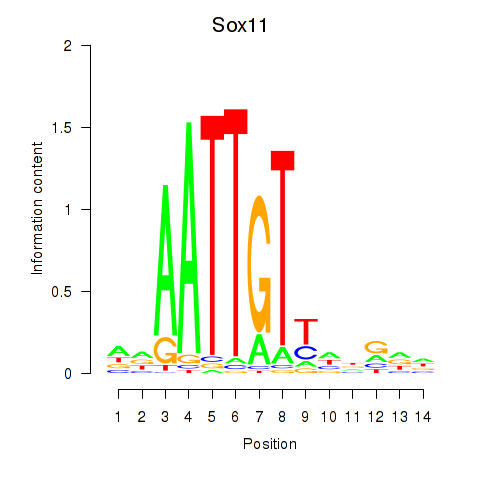
Results for Sox11
Z-value: 0.59
Transcription factors associated with Sox11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox11
|
ENSRNOG00000030034 | SRY box 11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox11 | rn6_v1_chr6_-_46631983_46631983 | 0.42 | 2.1e-15 | Click! |
Activity profile of Sox11 motif
Sorted Z-values of Sox11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_50278842 | 25.32 |
ENSRNOT00000088950
|
Syt1
|
synaptotagmin 1 |
| chr4_-_16669368 | 20.52 |
ENSRNOT00000007608
|
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr12_-_10335499 | 18.83 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chr2_-_183210799 | 17.30 |
ENSRNOT00000085382
|
Trim2
|
tripartite motif-containing 2 |
| chr1_-_233140237 | 16.75 |
ENSRNOT00000083372
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr3_+_5709236 | 14.84 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr11_+_66316606 | 14.28 |
ENSRNOT00000041715
|
Stxbp5l
|
syntaxin binding protein 5-like |
| chr16_-_73152921 | 11.71 |
ENSRNOT00000048602
|
Zmat4
|
zinc finger, matrin type 4 |
| chr8_-_14880644 | 10.62 |
ENSRNOT00000015977
|
Fat3
|
FAT atypical cadherin 3 |
| chr14_+_88549947 | 10.59 |
ENSRNOT00000086177
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chrX_+_9956370 | 10.41 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr1_+_144070754 | 9.10 |
ENSRNOT00000079989
|
Sh3gl3
|
SH3 domain-containing GRB2-like 3 |
| chr1_-_124039196 | 8.53 |
ENSRNOT00000051883
|
Chrna7
|
cholinergic receptor nicotinic alpha 7 subunit |
| chr2_+_199796881 | 8.01 |
ENSRNOT00000024423
|
Fmo5
|
flavin containing monooxygenase 5 |
| chr13_+_89805962 | 7.99 |
ENSRNOT00000074035
|
Tstd1
|
thiosulfate sulfurtransferase like domain containing 1 |
| chr20_+_41266566 | 7.94 |
ENSRNOT00000000653
|
Frk
|
fyn-related Src family tyrosine kinase |
| chr17_-_43537293 | 7.26 |
ENSRNOT00000091749
|
Slc17a3
|
solute carrier family 17 member 3 |
| chr13_+_57243877 | 6.94 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr8_+_106816152 | 6.91 |
ENSRNOT00000047613
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr4_-_11497531 | 6.35 |
ENSRNOT00000078799
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chrX_+_10042893 | 6.10 |
ENSRNOT00000084218
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr14_+_8080275 | 6.04 |
ENSRNOT00000065965
ENSRNOT00000092542 |
Mapk10
|
mitogen activated protein kinase 10 |
| chr14_+_8080565 | 5.85 |
ENSRNOT00000092395
|
Mapk10
|
mitogen activated protein kinase 10 |
| chr18_+_29993361 | 5.80 |
ENSRNOT00000075810
|
Pcdha4
|
protocadherin alpha 4 |
| chrX_+_65040934 | 5.79 |
ENSRNOT00000044006
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr2_-_259382765 | 5.72 |
ENSRNOT00000091407
|
St6galnac3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr3_-_66417741 | 5.67 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr2_+_22950018 | 5.61 |
ENSRNOT00000071804
|
Homer1
|
homer scaffolding protein 1 |
| chr9_+_95161157 | 5.57 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chrX_+_65040775 | 5.42 |
ENSRNOT00000081354
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr11_-_1983513 | 5.21 |
ENSRNOT00000000907
|
Htr1f
|
5-hydroxytryptamine receptor 1F |
| chr16_+_50152008 | 5.01 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr8_+_107229832 | 4.74 |
ENSRNOT00000045821
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr19_-_17494516 | 4.44 |
ENSRNOT00000078786
ENSRNOT00000076138 |
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr14_+_22072024 | 4.04 |
ENSRNOT00000002680
|
ste2
|
estrogen sulfotransferase |
| chr4_+_154215250 | 3.95 |
ENSRNOT00000072465
|
Mug2
|
murinoglobulin 2 |
| chr7_+_140829076 | 3.70 |
ENSRNOT00000086179
|
Spats2
|
spermatogenesis associated, serine-rich 2 |
| chr5_+_2278357 | 3.69 |
ENSRNOT00000066590
|
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr20_-_45053640 | 3.62 |
ENSRNOT00000072256
|
RGD1561777
|
similar to Na+ dependent glucose transporter 1 |
| chr10_-_58771908 | 3.46 |
ENSRNOT00000018808
|
RGD1304728
|
similar to 4933427D14Rik protein |
| chr4_+_22082194 | 3.34 |
ENSRNOT00000091799
|
Crot
|
carnitine O-octanoyltransferase |
| chr14_+_23507628 | 3.32 |
ENSRNOT00000037509
|
Uba6
|
ubiquitin-like modifier activating enzyme 6 |
| chr6_+_100337226 | 3.20 |
ENSRNOT00000011220
|
Fut8
|
fucosyltransferase 8 |
| chr4_+_31333970 | 3.08 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr1_+_207887209 | 3.07 |
ENSRNOT00000021359
|
Ptpre
|
protein tyrosine phosphatase, receptor type, E |
| chr10_-_49149479 | 3.02 |
ENSRNOT00000004291
|
Zfp287
|
zinc finger protein 287 |
| chr7_-_144837395 | 3.00 |
ENSRNOT00000089024
|
Cbx5
|
chromobox 5 |
| chr6_+_107603580 | 2.91 |
ENSRNOT00000036430
|
Dnal1
|
dynein, axonemal, light chain 1 |
| chr15_-_93748742 | 2.80 |
ENSRNOT00000093370
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr12_-_12546111 | 2.78 |
ENSRNOT00000046381
|
Lmtk2
|
lemur tyrosine kinase 2 |
| chr17_-_47653989 | 2.65 |
ENSRNOT00000072833
|
LOC498759
|
LRRGT00094 |
| chr14_+_62309299 | 2.49 |
ENSRNOT00000048309
|
Vom1r26
|
vomeronasal 1 receptor 26 |
| chr5_-_166726794 | 2.49 |
ENSRNOT00000022799
|
Slc25a33
|
solute carrier family 25 member 33 |
| chr10_-_52290657 | 2.43 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr2_+_56052643 | 2.40 |
ENSRNOT00000029565
|
Rictor
|
RPTOR independent companion of MTOR, complex 2 |
| chr7_-_17520780 | 2.30 |
ENSRNOT00000029672
|
RGD1560291
|
similar to NACHT, leucine rich repeat and PYD containing 4A |
| chr6_-_42616548 | 2.30 |
ENSRNOT00000081433
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr1_+_171793782 | 2.18 |
ENSRNOT00000081351
|
Olfml1
|
olfactomedin-like 1 |
| chr7_-_119689938 | 2.16 |
ENSRNOT00000000200
|
Tmprss6
|
transmembrane protease, serine 6 |
| chr5_-_147584038 | 2.15 |
ENSRNOT00000010983
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chrX_-_29825439 | 2.10 |
ENSRNOT00000048155
|
Gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr13_-_82295123 | 2.09 |
ENSRNOT00000090120
|
LOC498265
|
similar to hypothetical protein FLJ10706 |
| chr6_-_76552559 | 1.84 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr5_-_160070916 | 1.78 |
ENSRNOT00000067517
ENSRNOT00000055791 |
Spen
|
spen family transcriptional repressor |
| chr5_+_154077944 | 1.74 |
ENSRNOT00000031512
|
Il22ra1
|
interleukin 22 receptor subunit alpha 1 |
| chr3_+_80468154 | 1.68 |
ENSRNOT00000021837
|
Ckap5
|
cytoskeleton associated protein 5 |
| chr15_-_61876430 | 1.61 |
ENSRNOT00000049690
|
AC123280.1
|
|
| chr13_-_105684374 | 1.54 |
ENSRNOT00000073142
|
Spata17
|
spermatogenesis associated 17 |
| chr14_-_45165207 | 1.43 |
ENSRNOT00000002960
|
Klf3
|
Kruppel like factor 3 |
| chr2_+_206454208 | 1.42 |
ENSRNOT00000026863
|
Phtf1
|
putative homeodomain transcription factor 1 |
| chr5_-_117754571 | 1.38 |
ENSRNOT00000011405
|
Dock7
|
dedicator of cytokinesis 7 |
| chrX_+_63343546 | 1.34 |
ENSRNOT00000076315
|
Klhl15
|
kelch-like family member 15 |
| chr3_-_100364400 | 1.31 |
ENSRNOT00000006670
|
Mettl15
|
methyltransferase like 15 |
| chr1_-_124038966 | 1.28 |
ENSRNOT00000082487
|
Chrna7
|
cholinergic receptor nicotinic alpha 7 subunit |
| chr20_-_28783589 | 1.23 |
ENSRNOT00000071663
|
Sept10
|
septin 10 |
| chr7_+_65616177 | 1.20 |
ENSRNOT00000006566
|
Gns
|
glucosamine (N-acetyl)-6-sulfatase |
| chr5_+_171274544 | 1.17 |
ENSRNOT00000077307
|
Cep104
|
centrosomal protein 104 |
| chr17_-_2705123 | 1.06 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr18_-_26656879 | 0.90 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr5_-_144996431 | 0.89 |
ENSRNOT00000016992
|
Zmym4
|
zinc finger MYM-type containing 4 |
| chr15_-_108120279 | 0.86 |
ENSRNOT00000090572
|
Dock9
|
dedicator of cytokinesis 9 |
| chr6_+_137924568 | 0.83 |
ENSRNOT00000046456
|
Mta1
|
metastasis associated 1 |
| chr9_+_121456623 | 0.53 |
ENSRNOT00000056255
|
Ppidl1
|
peptidylprolyl isomerase D-like 1 |
| chr3_-_103485806 | 0.50 |
ENSRNOT00000072524
|
Olr789
|
olfactory receptor 789 |
| chr3_-_152179193 | 0.40 |
ENSRNOT00000026700
|
Rbm12
|
RNA binding motif protein 12 |
| chr3_+_102658573 | 0.36 |
ENSRNOT00000006545
|
Olr758
|
olfactory receptor 758 |
| chr19_-_29802083 | 0.34 |
ENSRNOT00000024755
|
AC123471.1
|
|
| chr3_+_75465399 | 0.31 |
ENSRNOT00000057479
|
Olr562
|
olfactory receptor 562 |
| chr3_-_78045379 | 0.30 |
ENSRNOT00000078664
|
Olr689
|
olfactory receptor 689 |
| chr19_+_39543242 | 0.28 |
ENSRNOT00000018837
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr6_+_22696397 | 0.14 |
ENSRNOT00000011630
|
Alk
|
anaplastic lymphoma receptor tyrosine kinase |
| chr7_-_13588411 | 0.10 |
ENSRNOT00000088783
|
Olr1083
|
olfactory receptor 1083 |
| chr5_+_35991068 | 0.07 |
ENSRNOT00000061139
|
Pnisr
|
PNN interacting serine and arginine rich protein |
| chr1_-_235405831 | 0.04 |
ENSRNOT00000071578
|
AABR07006458.1
|
|
| chr2_+_4989295 | 0.02 |
ENSRNOT00000041541
|
Fam172a
|
family with sequence similarity 172, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.8 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 3.4 | 20.5 | GO:0099526 | synaptic vesicle targeting(GO:0016080) presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 3.3 | 16.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 3.3 | 9.8 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 1.8 | 10.6 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 1.7 | 25.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 1.1 | 9.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.1 | 5.7 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 1.1 | 11.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.0 | 18.8 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.9 | 3.7 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.8 | 3.2 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.8 | 6.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.7 | 5.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.7 | 3.3 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.7 | 7.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.6 | 5.6 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.6 | 2.4 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.4 | 2.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.4 | 10.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.4 | 5.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.3 | 5.7 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.3 | 3.3 | GO:0021764 | amygdala development(GO:0021764) |
| 0.3 | 11.7 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.3 | 2.5 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.3 | 2.8 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.3 | 1.2 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.3 | 1.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 0.8 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.3 | 1.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.2 | 14.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.2 | 2.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 2.8 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.2 | 1.4 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 3.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 7.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 3.1 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.1 | 5.2 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 2.3 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 2.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0090648 | response to environmental enrichment(GO:0090648) |
| 0.0 | 14.1 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 5.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 3.0 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 7.2 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.5 | GO:0044317 | rod spherule(GO:0044317) |
| 6.7 | 40.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 3.5 | 10.6 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.6 | 6.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.5 | 9.8 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.5 | 2.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 6.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 2.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 2.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 3.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 5.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 9.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 3.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 8.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 6.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 11.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 14.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 6.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 5.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 7.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 7.4 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 5.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 8.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 25.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 4.9 | 14.8 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 1.6 | 6.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.3 | 10.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 1.2 | 11.9 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.1 | 5.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.1 | 20.5 | GO:0005522 | profilin binding(GO:0005522) |
| 1.1 | 5.6 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 1.1 | 3.3 | GO:0016414 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 1.1 | 3.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.7 | 7.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) efflux transmembrane transporter activity(GO:0015562) salt transmembrane transporter activity(GO:1901702) |
| 0.7 | 8.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.6 | 16.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.6 | 1.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.6 | 9.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.4 | 5.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.3 | 1.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 3.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.3 | 2.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 6.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 2.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 5.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 7.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 5.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 14.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 2.5 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 6.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 2.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 16.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 1.2 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.1 | 3.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 7.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 3.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 13.0 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 39.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 11.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 10.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 4.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 25.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.9 | 16.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.9 | 14.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.8 | 9.8 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.7 | 11.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 5.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 5.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 5.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 5.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 6.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 5.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 3.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 2.4 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.2 | 3.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 2.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 2.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 10.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 3.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |