Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
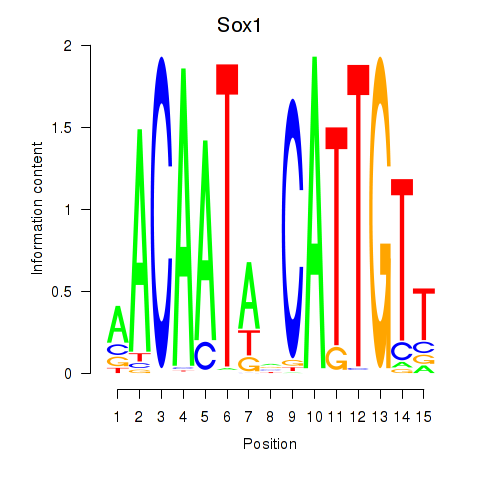
Results for Sox1
Z-value: 0.90
Transcription factors associated with Sox1
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Sox1 motif
Sorted Z-values of Sox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_81534140 | 49.65 |
ENSRNOT00000027202
|
Tex101
|
testis expressed 101 |
| chr10_-_5533695 | 44.12 |
ENSRNOT00000051564
|
Rpl39l
|
ribosomal protein L39-like |
| chr2_+_138194136 | 39.75 |
ENSRNOT00000014928
|
RGD1307595
|
similar to RIKEN cDNA 1700018B24 |
| chr18_-_41899450 | 39.62 |
ENSRNOT00000057099
|
RGD1561114
|
similar to hypothetical protein 4930474N05 |
| chr7_+_34744483 | 37.96 |
ENSRNOT00000007465
|
Usp44
|
ubiquitin specific peptidase 44 |
| chr7_+_34402738 | 36.65 |
ENSRNOT00000030985
|
Ccdc38
|
coiled-coil domain containing 38 |
| chr16_+_12174370 | 34.45 |
ENSRNOT00000072045
|
LOC102553538
|
ral guanine nucleotide dissociation stimulator-like |
| chr16_+_12976906 | 33.20 |
ENSRNOT00000029573
|
LOC685463
|
similar to Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS) |
| chr12_+_45319501 | 33.07 |
ENSRNOT00000090630
ENSRNOT00000041732 |
RGD1561114
|
similar to hypothetical protein 4930474N05 |
| chr16_-_12703961 | 32.53 |
ENSRNOT00000080905
|
RGD1562508
|
similar to hypothetical protein 4930474N05 |
| chr16_-_12194118 | 32.33 |
ENSRNOT00000071517
|
RGD1559804
|
similar to hypothetical protein 4930474N05 |
| chr2_-_228665279 | 30.74 |
ENSRNOT00000055648
|
RGD1306519
|
similar to T-cell activation Rho GTPase-activating protein isoform b |
| chr7_+_18310624 | 30.46 |
ENSRNOT00000075258
|
Actl9
|
actin-like 9 |
| chr16_-_12124265 | 29.92 |
ENSRNOT00000080192
|
AABR07024682.1
|
|
| chr16_-_12742010 | 29.58 |
ENSRNOT00000077513
|
LOC100911649
|
ral guanine nucleotide dissociation stimulator-like |
| chr3_+_148082790 | 29.33 |
ENSRNOT00000009930
ENSRNOT00000080767 |
Defb36
|
defensin beta 36 |
| chr10_-_41491554 | 29.17 |
ENSRNOT00000035759
|
LOC102552619
|
uncharacterized LOC102552619 |
| chr20_+_4959294 | 29.17 |
ENSRNOT00000074223
|
Hspa1l
|
heat shock protein family A (Hsp70) member 1 like |
| chr19_+_14835822 | 25.94 |
ENSRNOT00000072804
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene |
| chr16_-_12538117 | 25.27 |
ENSRNOT00000046081
|
AABR07024718.1
|
|
| chr19_-_36157924 | 24.50 |
ENSRNOT00000072022
|
AABR07043701.1
|
|
| chr16_+_13050305 | 24.37 |
ENSRNOT00000077388
|
AABR07024735.1
|
|
| chr16_+_12510827 | 24.19 |
ENSRNOT00000077763
|
AABR07024716.1
|
|
| chr16_-_12626949 | 23.96 |
ENSRNOT00000077469
|
AABR07024722.1
|
|
| chr16_-_12662336 | 23.76 |
ENSRNOT00000080161
|
RGD1559508
|
similar to hypothetical protein 4930474N05 |
| chrX_+_35869538 | 23.39 |
ENSRNOT00000058947
|
Ppef1
|
protein phosphatase with EF-hand domain 1 |
| chr3_+_121128368 | 17.43 |
ENSRNOT00000055882
|
LOC499886
|
similar to hypothetical protein 4933411G11 |
| chr10_+_53740841 | 17.40 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr17_-_15030364 | 17.09 |
ENSRNOT00000044543
|
Nutm2f
|
NUT family member 2F |
| chr4_+_70252366 | 16.62 |
ENSRNOT00000073039
|
Chl1
|
cell adhesion molecule L1-like |
| chr10_+_16635989 | 14.94 |
ENSRNOT00000028155
|
Nkx2-5
|
NK2 homeobox 5 |
| chr1_+_142136452 | 14.14 |
ENSRNOT00000016445
|
Hddc3
|
HD domain containing 3 |
| chr1_+_199449973 | 12.13 |
ENSRNOT00000029994
|
Trim72
|
tripartite motif containing 72 |
| chr1_+_148240504 | 11.50 |
ENSRNOT00000085373
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr1_-_224533219 | 11.09 |
ENSRNOT00000051289
|
Ust5r
|
integral membrane transport protein UST5r |
| chr20_+_6869767 | 10.35 |
ENSRNOT00000000631
ENSRNOT00000086330 ENSRNOT00000093188 ENSRNOT00000093736 |
RGD735065
|
similar to GI:13385412-like protein splice form I |
| chr14_-_21553949 | 9.41 |
ENSRNOT00000031642
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chr4_-_176231344 | 9.12 |
ENSRNOT00000049154
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr18_+_52215682 | 8.86 |
ENSRNOT00000037901
|
Megf10
|
multiple EGF-like domains 10 |
| chr20_+_27129291 | 8.71 |
ENSRNOT00000031053
|
AABR07044933.1
|
|
| chr11_+_65960277 | 8.49 |
ENSRNOT00000003674
|
Ndufb4
|
NADH:ubiquinone oxidoreductase subunit B4 |
| chr14_-_21615488 | 7.90 |
ENSRNOT00000002667
|
Vcsa2
|
variable coding sequence A2 |
| chr2_+_157453428 | 7.51 |
ENSRNOT00000051494
|
Lekr1
|
leucine, glutamate and lysine rich 1 |
| chr3_+_63510293 | 5.79 |
ENSRNOT00000058093
|
Dfnb59
|
deafness, autosomal recessive 59 |
| chr17_-_14637832 | 5.62 |
ENSRNOT00000074323
|
LOC103690116
|
osteomodulin |
| chr20_-_556982 | 5.15 |
ENSRNOT00000047478
|
Olr1683
|
olfactory receptor 1683 |
| chr13_-_61306939 | 4.58 |
ENSRNOT00000086610
|
Rgs21
|
regulator of G-protein signaling 21 |
| chr9_-_70374553 | 4.53 |
ENSRNOT00000064447
|
Dytn
|
dystrotelin |
| chr15_-_27408258 | 4.17 |
ENSRNOT00000043588
|
RGD1562558
|
similar to Cytochrome c, somatic |
| chr19_+_22699808 | 4.09 |
ENSRNOT00000023169
|
RGD1308706
|
similar to RIKEN cDNA 4921524J17 |
| chr4_-_170810080 | 3.97 |
ENSRNOT00000082312
|
Wbp11
|
WW domain binding protein 11 |
| chr1_+_141120166 | 3.84 |
ENSRNOT00000050759
|
Fanci
|
Fanconi anemia, complementation group I |
| chr8_+_43705216 | 3.68 |
ENSRNOT00000090425
|
AC115384.1
|
|
| chr4_-_170783725 | 3.56 |
ENSRNOT00000072528
|
Wbp11
|
WW domain binding protein 11 |
| chr3_-_10371240 | 3.49 |
ENSRNOT00000012075
|
Ass1
|
argininosuccinate synthase 1 |
| chr15_+_49069145 | 3.27 |
ENSRNOT00000019487
|
Scara5
|
scavenger receptor class A, member 5 |
| chr7_+_91833297 | 3.16 |
ENSRNOT00000079792
|
Slc30a8
|
solute carrier family 30 member 8 |
| chr4_-_2201749 | 3.02 |
ENSRNOT00000089327
|
Lmbr1
|
limb development membrane protein 1 |
| chr3_+_102703487 | 3.01 |
ENSRNOT00000089603
|
AC121203.2
|
|
| chr3_+_102664391 | 2.80 |
ENSRNOT00000041791
|
RGD1561276
|
similar to olfactory receptor Olfr1289 |
| chr1_+_170796271 | 2.55 |
ENSRNOT00000042820
|
Olr210
|
olfactory receptor 210 |
| chr7_+_101146820 | 2.47 |
ENSRNOT00000080206
|
AABR07058124.3
|
|
| chr8_+_23771920 | 2.44 |
ENSRNOT00000020481
|
Bbs9
|
Bardet-Biedl syndrome 9 |
| chr17_+_66446569 | 2.32 |
ENSRNOT00000070825
|
Heatr1
|
HEAT repeat containing 1 |
| chr17_-_15478343 | 2.29 |
ENSRNOT00000078029
|
Omd
|
osteomodulin |
| chr20_+_1124169 | 2.15 |
ENSRNOT00000072544
|
LOC100910894
|
olfactory receptor 14J1-like |
| chr7_-_12569110 | 2.03 |
ENSRNOT00000093446
ENSRNOT00000016733 |
Wdr18
|
WD repeat domain 18 |
| chr7_+_144080614 | 2.02 |
ENSRNOT00000077687
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr20_+_1012054 | 1.96 |
ENSRNOT00000073732
|
Olr1704
|
olfactory receptor 1704 |
| chr3_+_102723402 | 1.67 |
ENSRNOT00000040298
|
Olr765
|
olfactory receptor 765 |
| chr2_+_193483793 | 1.34 |
ENSRNOT00000074518
|
Flg2
|
filaggrin family member 2 |
| chr10_-_84920886 | 0.93 |
ENSRNOT00000068083
|
Sp2
|
Sp2 transcription factor |
| chr18_-_62476700 | 0.70 |
ENSRNOT00000048429
|
Cycs
|
cytochrome c, somatic |
| chr13_-_84694575 | 0.37 |
ENSRNOT00000066609
|
Fmo13
|
flavin-containing monooxygenase 13 |
| chr10_-_109827143 | 0.20 |
ENSRNOT00000054948
|
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr10_+_39066716 | 0.11 |
ENSRNOT00000010729
|
Il5
|
interleukin 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 14.9 | GO:0003162 | atrioventricular node development(GO:0003162) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) septum secundum development(GO:0003285) His-Purkinje system cell differentiation(GO:0060932) |
| 2.7 | 38.0 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 2.3 | 29.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 1.3 | 8.9 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) skeletal muscle satellite cell differentiation(GO:0014816) recognition of apoptotic cell(GO:0043654) |
| 1.2 | 39.8 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 1.2 | 29.2 | GO:0042026 | protein refolding(GO:0042026) |
| 1.2 | 3.5 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 1.1 | 49.7 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 1.0 | 10.3 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.4 | 9.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.4 | 11.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.4 | 7.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.4 | 16.6 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.3 | 162.1 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.3 | 3.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 2.0 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.2 | 2.0 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 0.7 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 3.8 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 3.3 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 7.9 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.1 | 44.1 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 1.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 2.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 14.3 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 3.0 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 15.5 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 5.8 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 9.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.9 | GO:0048144 | fibroblast proliferation(GO:0048144) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 29.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 1.9 | 17.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.8 | 49.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.4 | 44.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 2.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.3 | 8.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 2.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 3.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 8.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 36.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 9.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 12.1 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 16.6 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 3.2 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 6.1 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.7 | 168.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.5 | 38.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.4 | 2.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.4 | 11.5 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.4 | 23.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.3 | 8.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 12.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.3 | 29.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 7.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 3.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 14.9 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.2 | 3.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 44.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 3.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 16.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 3.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 9.4 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.7 | GO:0009055 | electron carrier activity(GO:0009055) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 17.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 15.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 3.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 10.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 2.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 9.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 3.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |