Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
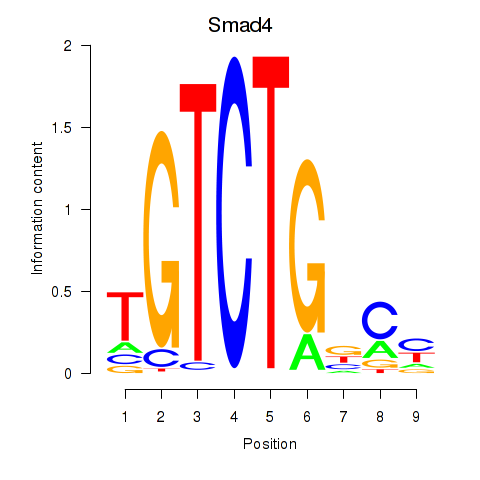
Results for Smad4
Z-value: 1.00
Transcription factors associated with Smad4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad4
|
ENSRNOG00000051965 | SMAD family member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad4 | rn6_v1_chr18_-_69657301_69657301 | 0.46 | 5.7e-18 | Click! |
Activity profile of Smad4 motif
Sorted Z-values of Smad4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_56862691 | 39.59 |
ENSRNOT00000087712
|
Gad1
|
glutamate decarboxylase 1 |
| chr16_-_39476384 | 28.64 |
ENSRNOT00000092968
|
Gpm6a
|
glycoprotein m6a |
| chr13_-_91872954 | 27.80 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr10_-_45297385 | 26.55 |
ENSRNOT00000041187
|
Hist3h2bb
|
histone cluster 3 H2B family member b |
| chr10_-_107539658 | 25.72 |
ENSRNOT00000089346
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr1_+_196996581 | 25.64 |
ENSRNOT00000021690
|
Il21r
|
interleukin 21 receptor |
| chr2_+_209839299 | 25.56 |
ENSRNOT00000092450
|
Kcna2
|
potassium voltage-gated channel subfamily A member 2 |
| chr2_+_209838869 | 25.51 |
ENSRNOT00000092365
|
Kcna2
|
potassium voltage-gated channel subfamily A member 2 |
| chr16_-_39476025 | 25.16 |
ENSRNOT00000014312
|
Gpm6a
|
glycoprotein m6a |
| chr10_-_107539465 | 25.11 |
ENSRNOT00000004524
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr17_-_43770561 | 22.62 |
ENSRNOT00000088408
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr8_+_58347736 | 21.49 |
ENSRNOT00000080227
ENSRNOT00000066222 |
Slc35f2
|
solute carrier family 35, member F2 |
| chr1_+_100299626 | 21.27 |
ENSRNOT00000092327
ENSRNOT00000044257 |
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr8_-_98738446 | 21.25 |
ENSRNOT00000019860
|
Zic1
|
Zic family member 1 |
| chr4_+_153774486 | 20.63 |
ENSRNOT00000074096
|
Tuba8
|
tubulin, alpha 8 |
| chr1_+_78800754 | 20.55 |
ENSRNOT00000084601
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr17_-_44744902 | 17.41 |
ENSRNOT00000085381
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr10_-_94352880 | 17.28 |
ENSRNOT00000035973
|
Limd2
|
LIM domain containing 2 |
| chr9_+_117583610 | 17.06 |
ENSRNOT00000088647
ENSRNOT00000049426 |
Epb41l3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr7_-_120403523 | 16.69 |
ENSRNOT00000015283
|
Sox10
|
SRY box 10 |
| chr17_+_44520537 | 16.30 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr7_-_70842405 | 16.20 |
ENSRNOT00000047449
|
Nxph4
|
neurexophilin 4 |
| chr9_+_2190915 | 15.81 |
ENSRNOT00000077417
|
Satb1
|
SATB homeobox 1 |
| chr18_+_52215682 | 14.55 |
ENSRNOT00000037901
|
Megf10
|
multiple EGF-like domains 10 |
| chr8_-_50228369 | 14.39 |
ENSRNOT00000024030
|
Tagln
|
transgelin |
| chr13_-_70625842 | 13.87 |
ENSRNOT00000092499
|
Lamc2
|
laminin subunit gamma 2 |
| chr5_-_152473868 | 12.98 |
ENSRNOT00000022130
|
Fam110d
|
family with sequence similarity 110, member D |
| chr17_-_43821536 | 12.50 |
ENSRNOT00000072286
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr13_-_70626252 | 12.47 |
ENSRNOT00000036947
|
Lamc2
|
laminin subunit gamma 2 |
| chr17_-_44520240 | 12.31 |
ENSRNOT00000086538
|
Hist1h2bh
|
histone cluster 1 H2B family member h |
| chr1_-_72727112 | 11.38 |
ENSRNOT00000031172
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr1_+_99486253 | 11.32 |
ENSRNOT00000074820
|
LOC102546648
|
uncharacterized LOC102546648 |
| chr7_+_119647375 | 11.07 |
ENSRNOT00000046563
|
Kctd17
|
potassium channel tetramerization domain containing 17 |
| chr7_+_123168811 | 10.20 |
ENSRNOT00000007091
|
Csdc2
|
cold shock domain containing C2 |
| chr12_+_21767606 | 10.07 |
ENSRNOT00000059602
|
LOC103691013
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr14_-_34561696 | 9.79 |
ENSRNOT00000059763
|
Srd5a3
|
steroid 5 alpha-reductase 3 |
| chr13_+_57131395 | 9.66 |
ENSRNOT00000017884
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr16_+_61758917 | 8.82 |
ENSRNOT00000084858
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chrX_+_106823491 | 8.77 |
ENSRNOT00000045997
|
Bex3
|
brain expressed X-linked 3 |
| chr15_+_4240203 | 8.59 |
ENSRNOT00000010178
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr13_+_48287873 | 8.51 |
ENSRNOT00000068223
|
Fam72a
|
family with sequence similarity 72, member A |
| chr4_+_102351036 | 8.48 |
ENSRNOT00000079277
|
AABR07060994.1
|
|
| chr4_-_170145262 | 8.39 |
ENSRNOT00000074526
|
Hist1h4b
|
histone cluster 1, H4b |
| chr2_+_241909832 | 7.82 |
ENSRNOT00000047975
|
Ppp3ca
|
protein phosphatase 3 catalytic subunit alpha |
| chrX_+_96991658 | 7.80 |
ENSRNOT00000049969
|
AABR07040288.1
|
|
| chr2_-_187822997 | 7.78 |
ENSRNOT00000092932
|
Sema4a
|
semaphorin 4A |
| chr10_-_90240509 | 7.61 |
ENSRNOT00000028407
|
Atxn7l3
|
ataxin 7-like 3 |
| chr17_-_9695292 | 7.28 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr7_-_14418950 | 7.01 |
ENSRNOT00000066722
|
Rasal3
|
RAS protein activator like 3 |
| chr1_+_79989019 | 6.85 |
ENSRNOT00000020428
|
Dmpk
|
dystrophia myotonica-protein kinase |
| chr8_-_115358046 | 6.81 |
ENSRNOT00000017607
|
Grm2
|
glutamate metabotropic receptor 2 |
| chr4_-_103369224 | 6.75 |
ENSRNOT00000075709
|
AABR07061057.1
|
|
| chr10_+_47765432 | 6.63 |
ENSRNOT00000078231
|
LOC102553715
|
microfibril-associated glycoprotein 4-like |
| chrX_+_107370431 | 5.47 |
ENSRNOT00000044372
|
Tceal1
|
transcription elongation factor A like 1 |
| chr13_-_94355219 | 5.29 |
ENSRNOT00000005332
|
Pld5
|
phospholipase D family, member 5 |
| chr15_+_23792931 | 5.24 |
ENSRNOT00000092091
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr1_-_101514547 | 4.99 |
ENSRNOT00000079633
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr15_-_29852665 | 4.99 |
ENSRNOT00000040046
|
AABR07017662.3
|
|
| chr2_+_198360998 | 4.92 |
ENSRNOT00000046129
|
Hist2h2be
|
histone cluster 2, H2be |
| chr10_+_68588789 | 4.89 |
ENSRNOT00000049614
|
AABR07030122.1
|
|
| chr17_+_78739389 | 4.79 |
ENSRNOT00000020384
|
LOC102547897
|
uncharacterized LOC102547897 |
| chr2_+_203768729 | 4.78 |
ENSRNOT00000018895
|
Igsf3
|
immunoglobulin superfamily, member 3 |
| chr4_-_82209933 | 4.54 |
ENSRNOT00000091106
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr4_-_82295470 | 4.48 |
ENSRNOT00000091073
|
Hoxa10
|
homeobox A10 |
| chr4_+_84194347 | 4.38 |
ENSRNOT00000084256
|
Chn2
|
chimerin 2 |
| chr14_-_35149608 | 3.85 |
ENSRNOT00000003050
ENSRNOT00000090654 |
Kit
|
KIT proto-oncogene receptor tyrosine kinase |
| chr2_-_5577369 | 3.71 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr10_-_88670430 | 3.58 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr6_+_108745895 | 3.26 |
ENSRNOT00000061786
|
LOC108348062
|
eukaryotic translation initiation factor 3 subunit H |
| chr12_+_37668369 | 3.15 |
ENSRNOT00000001419
|
Cdk2ap1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr17_+_23135985 | 2.12 |
ENSRNOT00000090794
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr5_-_152464850 | 2.05 |
ENSRNOT00000021937
|
Zfp593
|
zinc finger protein 593 |
| chr15_-_29865536 | 2.01 |
ENSRNOT00000085893
|
AABR07017658.1
|
|
| chr1_-_168587241 | 1.83 |
ENSRNOT00000021254
|
Olr104
|
olfactory receptor 104 |
| chr8_+_36766977 | 1.55 |
ENSRNOT00000017648
|
Pus3
|
pseudouridylate synthase 3 |
| chr10_+_46940965 | 1.51 |
ENSRNOT00000005366
|
Llgl1
|
LLGL1, scribble cell polarity complex component |
| chr7_-_143217535 | 1.39 |
ENSRNOT00000088316
|
Kb15
|
type II keratin Kb15 |
| chr12_-_41671437 | 1.29 |
ENSRNOT00000001883
|
Lhx5
|
LIM homeobox 5 |
| chr3_+_102683148 | 0.77 |
ENSRNOT00000046220
|
RGD1566059
|
similar to olfactory receptor Olfr1289 |
| chr6_+_29012207 | 0.68 |
ENSRNOT00000079077
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr10_+_56453877 | 0.63 |
ENSRNOT00000031640
|
Plscr3
|
phospholipid scramblase 3 |
| chr12_+_4546287 | 0.50 |
ENSRNOT00000001416
|
Higd2al1
|
HIG1 hypoxia inducible domain family, member 2A-like 1 |
| chr15_+_32188736 | 0.39 |
ENSRNOT00000081436
|
AABR07017878.1
|
|
| chr8_+_117280705 | 0.27 |
ENSRNOT00000085038
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr11_+_9642365 | 0.16 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr3_+_172155496 | 0.03 |
ENSRNOT00000066279
|
Stx16
|
syntaxin 16 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Smad4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 51.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 7.9 | 39.6 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 5.3 | 21.3 | GO:0050893 | sensory processing(GO:0050893) |
| 3.6 | 60.9 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 2.8 | 17.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 2.1 | 14.5 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) skeletal muscle satellite cell differentiation(GO:0014816) recognition of apoptotic cell(GO:0043654) |
| 2.0 | 9.8 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 2.0 | 7.8 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 1.8 | 7.0 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 1.7 | 5.0 | GO:1902310 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) response to environmental enrichment(GO:0090648) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 1.7 | 21.5 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 1.5 | 20.6 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 1.4 | 6.9 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 1.3 | 8.8 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 1.3 | 26.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 1.3 | 53.8 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 1.1 | 15.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 1.1 | 8.5 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 1.0 | 50.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 1.0 | 3.8 | GO:1905065 | Kit signaling pathway(GO:0038109) melanocyte migration(GO:0097324) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.6 | 6.8 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.5 | 27.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.5 | 3.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.4 | 3.6 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.4 | 21.3 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.4 | 11.1 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.4 | 7.8 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.4 | 1.6 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.4 | 34.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.4 | 7.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.4 | 4.9 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.3 | 4.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.3 | 2.0 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 5.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 16.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.2 | 1.3 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.2 | 3.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 11.4 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.1 | 8.6 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 10.2 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 4.5 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 1.5 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.1 | 8.8 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 3.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 25.8 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 9.7 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 26.3 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 4.3 | 68.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 3.3 | 16.7 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 2.2 | 60.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 1.3 | 7.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.2 | 53.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 1.0 | 39.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.8 | 13.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.8 | 55.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.6 | 7.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.5 | 14.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 7.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.4 | 3.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.4 | 5.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 15.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.3 | 50.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 6.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 27.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 1.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 3.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 8.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 9.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 6.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 11.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 5.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 7.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 3.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 8.8 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 8.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 4.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.9 | 39.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 3.0 | 21.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) ankyrin repeat binding(GO:0071532) |
| 2.6 | 51.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 2.4 | 9.8 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 2.3 | 6.8 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 2.2 | 8.8 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 2.1 | 14.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.0 | 7.8 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.6 | 20.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 1.5 | 4.4 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 0.9 | 11.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.7 | 2.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.7 | 11.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.5 | 3.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 3.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.4 | 53.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.4 | 9.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 37.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.3 | 61.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.3 | 66.3 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.2 | 25.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.2 | 16.7 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.2 | 5.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 3.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 26.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 7.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 14.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 3.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 6.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 55.1 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 20.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 15.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 3.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 17.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 8.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) LRR domain binding(GO:0030275) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 26.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 15.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 7.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 11.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 4.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 5.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 14.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 3.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 7.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 52.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 1.5 | 39.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 1.0 | 51.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.8 | 9.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 7.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 15.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.3 | 6.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 26.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.3 | 7.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 5.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 3.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 4.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |