Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Smad3
Z-value: 1.10
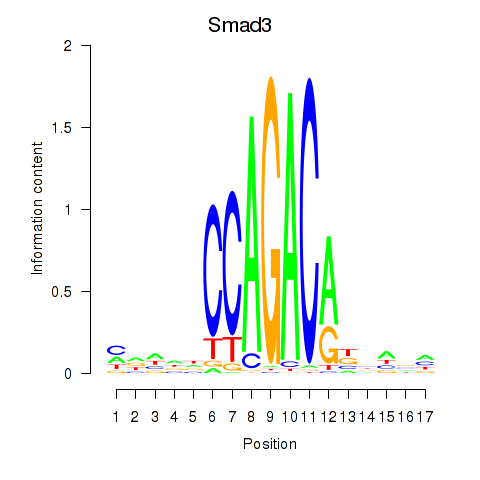
Transcription factors associated with Smad3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad3
|
ENSRNOG00000008620 | SMAD family member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad3 | rn6_v1_chr8_-_68678349_68678349 | 0.05 | 3.9e-01 | Click! |
Activity profile of Smad3 motif
Sorted Z-values of Smad3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_45297385 | 60.37 |
ENSRNOT00000041187
|
Hist3h2bb
|
histone cluster 3 H2B family member b |
| chr17_-_44748188 | 46.15 |
ENSRNOT00000081970
|
LOC103690190
|
histone H2A type 1-E |
| chr17_-_44841382 | 43.94 |
ENSRNOT00000080119
|
Hist1h2ak
|
histone cluster 1, H2ak |
| chr17_-_44815995 | 42.30 |
ENSRNOT00000091201
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr17_+_44738643 | 40.06 |
ENSRNOT00000087643
|
LOC100910554
|
histone H2A type 1-like |
| chr17_+_44748482 | 34.13 |
ENSRNOT00000083765
|
Hist1h2bl
|
histone cluster 1 H2B family member l |
| chr17_+_44528125 | 33.77 |
ENSRNOT00000084538
|
LOC680322
|
similar to Histone H2A type 1 |
| chr17_+_44520537 | 33.16 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr2_-_95472115 | 32.90 |
ENSRNOT00000015720
|
Stmn2
|
stathmin 2 |
| chr17_-_43770561 | 30.36 |
ENSRNOT00000088408
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr15_+_52767442 | 29.52 |
ENSRNOT00000014441
|
LOC108348161
|
phytanoyl-CoA hydroxylase-interacting protein |
| chr2_+_83393282 | 28.09 |
ENSRNOT00000044871
|
Ctnnd2
|
catenin delta 2 |
| chr4_-_170117325 | 25.19 |
ENSRNOT00000074198
|
LOC100912564
|
histone H4-like |
| chr3_-_94182714 | 24.99 |
ENSRNOT00000015073
|
LOC100362814
|
hypothetical protein LOC100362814 |
| chr17_-_44520240 | 24.90 |
ENSRNOT00000086538
|
Hist1h2bh
|
histone cluster 1 H2B family member h |
| chr1_+_192233910 | 24.86 |
ENSRNOT00000016418
ENSRNOT00000016442 |
Prkcb
|
protein kinase C, beta |
| chr15_+_52148379 | 24.48 |
ENSRNOT00000074912
|
Phyhip
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr17_+_44556039 | 23.85 |
ENSRNOT00000086540
|
Prss16
|
protease, serine 16 |
| chr17_-_44744902 | 22.76 |
ENSRNOT00000085381
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr17_-_43821536 | 21.64 |
ENSRNOT00000072286
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr13_+_90723092 | 20.68 |
ENSRNOT00000010146
|
Kcnj10
|
ATP-sensitive inward rectifier potassium channel 10 |
| chr17_-_44527801 | 20.43 |
ENSRNOT00000089643
|
Hist1h2bk
|
histone cluster 1 H2B family member k |
| chr7_+_123168811 | 20.22 |
ENSRNOT00000007091
|
Csdc2
|
cold shock domain containing C2 |
| chr9_-_89193821 | 20.17 |
ENSRNOT00000090881
|
Sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr8_+_58347736 | 18.88 |
ENSRNOT00000080227
ENSRNOT00000066222 |
Slc35f2
|
solute carrier family 35, member F2 |
| chr17_+_43673294 | 18.63 |
ENSRNOT00000074109
|
Hist2h4a
|
histone cluster 2, H4 |
| chr9_+_73319710 | 18.47 |
ENSRNOT00000092485
|
Map2
|
microtubule-associated protein 2 |
| chr1_+_72860218 | 18.19 |
ENSRNOT00000024547
|
Syt5
|
synaptotagmin 5 |
| chr1_-_226791773 | 18.14 |
ENSRNOT00000082482
ENSRNOT00000065376 ENSRNOT00000054812 ENSRNOT00000086669 |
LOC100911215
|
T-cell surface glycoprotein CD5-like |
| chr2_+_209839299 | 17.08 |
ENSRNOT00000092450
|
Kcna2
|
potassium voltage-gated channel subfamily A member 2 |
| chr5_+_122100099 | 16.81 |
ENSRNOT00000007738
|
Pde4b
|
phosphodiesterase 4B |
| chr2_+_209838869 | 16.03 |
ENSRNOT00000092365
|
Kcna2
|
potassium voltage-gated channel subfamily A member 2 |
| chr3_+_145032200 | 16.00 |
ENSRNOT00000068210
|
Syndig1
|
synapse differentiation inducing 1 |
| chr13_+_48287873 | 15.86 |
ENSRNOT00000068223
|
Fam72a
|
family with sequence similarity 72, member A |
| chr8_-_72204730 | 15.47 |
ENSRNOT00000023810
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr18_+_30398113 | 15.06 |
ENSRNOT00000027206
|
Pcdhb5
|
protocadherin beta 5 |
| chr14_-_33677031 | 14.86 |
ENSRNOT00000002908
|
RGD1311575
|
hypothetical LOC289568 |
| chr5_+_145257714 | 14.85 |
ENSRNOT00000019214
|
Dlgap3
|
DLG associated protein 3 |
| chr2_-_210282352 | 14.75 |
ENSRNOT00000075653
|
Slc6a17
|
solute carrier family 6 member 17 |
| chr1_-_84812486 | 13.98 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chrX_+_35869538 | 13.91 |
ENSRNOT00000058947
|
Ppef1
|
protein phosphatase with EF-hand domain 1 |
| chr7_+_123482255 | 13.69 |
ENSRNOT00000064487
|
LOC688613
|
hypothetical protein LOC688613 |
| chr13_+_99173484 | 13.10 |
ENSRNOT00000080574
ENSRNOT00000088654 |
Lefty2
|
Left-right determination factor 2 |
| chr18_-_69780922 | 12.82 |
ENSRNOT00000079093
|
Me2
|
malic enzyme 2 |
| chr7_-_116106368 | 12.60 |
ENSRNOT00000035678
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr2_+_198360998 | 12.27 |
ENSRNOT00000046129
|
Hist2h2be
|
histone cluster 2, H2be |
| chr7_-_119996824 | 12.08 |
ENSRNOT00000011079
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr10_-_90393317 | 11.76 |
ENSRNOT00000028563
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr3_-_80000091 | 11.71 |
ENSRNOT00000079394
|
Madd
|
MAP-kinase activating death domain |
| chr5_+_48456757 | 11.52 |
ENSRNOT00000091482
|
Srsf12
|
serine and arginine rich splicing factor 12 |
| chr9_-_65737538 | 11.35 |
ENSRNOT00000014939
|
Trak2
|
trafficking kinesin protein 2 |
| chr6_-_115513354 | 10.85 |
ENSRNOT00000005881
|
Ston2
|
stonin 2 |
| chr2_-_198360678 | 10.83 |
ENSRNOT00000051917
|
Hist2h2ac
|
histone cluster 2 H2A family member c |
| chr8_-_115358046 | 10.73 |
ENSRNOT00000017607
|
Grm2
|
glutamate metabotropic receptor 2 |
| chr1_-_101236065 | 10.33 |
ENSRNOT00000066834
|
Cd37
|
CD37 molecule |
| chrX_+_114929029 | 10.26 |
ENSRNOT00000006459
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr15_+_31395772 | 10.07 |
ENSRNOT00000075087
|
AABR07017824.1
|
|
| chr13_-_51076852 | 9.57 |
ENSRNOT00000078993
|
Adora1
|
adenosine A1 receptor |
| chr14_-_35149608 | 9.39 |
ENSRNOT00000003050
ENSRNOT00000090654 |
Kit
|
KIT proto-oncogene receptor tyrosine kinase |
| chr7_-_93280009 | 9.39 |
ENSRNOT00000064877
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr6_+_86823684 | 8.45 |
ENSRNOT00000086081
ENSRNOT00000006574 |
Fancm
|
Fanconi anemia, complementation group M |
| chr18_+_30474947 | 7.81 |
ENSRNOT00000027188
|
Pcdhb9
|
protocadherin beta 9 |
| chr3_+_152752091 | 7.68 |
ENSRNOT00000037177
|
Dlgap4
|
DLG associated protein 4 |
| chr1_-_165997751 | 7.67 |
ENSRNOT00000050227
|
P2ry6
|
pyrimidinergic receptor P2Y6 |
| chr8_-_72284852 | 7.27 |
ENSRNOT00000080319
ENSRNOT00000057739 |
Usp3
|
ubiquitin specific peptidase 3 |
| chr5_+_155935554 | 7.25 |
ENSRNOT00000031855
|
LOC100911993
|
ubiquitin carboxyl-terminal hydrolase 48-like |
| chr15_+_31456265 | 6.90 |
ENSRNOT00000075092
|
AABR07017825.1
|
|
| chr1_-_80514728 | 6.87 |
ENSRNOT00000081565
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chr10_+_106264434 | 6.75 |
ENSRNOT00000003891
|
Sept9
|
septin 9 |
| chr3_-_52510507 | 6.58 |
ENSRNOT00000091259
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr12_+_16950728 | 6.28 |
ENSRNOT00000060370
|
Ints1
|
integrator complex subunit 1 |
| chr8_+_117780891 | 5.92 |
ENSRNOT00000077236
|
Shisa5
|
shisa family member 5 |
| chr1_-_213987053 | 5.76 |
ENSRNOT00000072774
|
LOC100911519
|
p53-induced protein with a death domain-like |
| chr11_+_84972132 | 5.73 |
ENSRNOT00000034498
|
Lamp3
|
lysosomal-associated membrane protein 3 |
| chr3_-_112676556 | 5.57 |
ENSRNOT00000014664
|
Cdan1
|
codanin 1 |
| chr18_+_3861539 | 5.37 |
ENSRNOT00000015363
|
Lama3
|
laminin subunit alpha 3 |
| chrX_-_20807216 | 5.19 |
ENSRNOT00000075757
|
Fam156b
|
family with sequence similarity 156, member B |
| chr6_-_140880070 | 4.87 |
ENSRNOT00000073779
|
LOC691828
|
uncharacterized LOC691828 |
| chr8_-_46757781 | 4.78 |
ENSRNOT00000085294
|
AABR07073392.1
|
|
| chr10_+_45297937 | 4.61 |
ENSRNOT00000066367
|
Trim17
|
tripartite motif-containing 17 |
| chr18_+_64008048 | 4.54 |
ENSRNOT00000083414
|
Ldlrad4
|
low density lipoprotein receptor class A domain containing 4 |
| chr10_+_49541051 | 4.48 |
ENSRNOT00000041606
|
Pmp22
|
peripheral myelin protein 22 |
| chr11_-_84583542 | 4.24 |
ENSRNOT00000064212
|
Yeats2
|
YEATS domain containing 2 |
| chr1_+_259958310 | 4.17 |
ENSRNOT00000019751
|
LOC103689954
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr1_-_81152384 | 4.15 |
ENSRNOT00000026263
ENSRNOT00000077305 |
Zfp94
|
zinc finger protein 94 |
| chr18_+_30424814 | 4.09 |
ENSRNOT00000073425
|
Pcdhb7
|
protocadherin beta 7 |
| chr4_-_7228675 | 3.86 |
ENSRNOT00000064386
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr1_-_206417143 | 3.72 |
ENSRNOT00000091863
ENSRNOT00000073836 |
LOC100302465
|
hypothetical LOC100302465 |
| chr10_+_104483042 | 3.63 |
ENSRNOT00000007362
|
Sap30bp
|
SAP30 binding protein |
| chr16_+_84465656 | 3.60 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr12_+_18074033 | 3.56 |
ENSRNOT00000001727
|
LOC103689975
|
integrator complex subunit 1-like |
| chr7_+_16304954 | 3.56 |
ENSRNOT00000050751
|
Olr927
|
olfactory receptor 927 |
| chr2_-_12312287 | 3.55 |
ENSRNOT00000048860
|
AABR07007399.1
|
|
| chr7_+_115956276 | 3.53 |
ENSRNOT00000007773
|
Them6
|
thioesterase superfamily member 6 |
| chr3_-_111037425 | 3.40 |
ENSRNOT00000085628
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr15_+_31051414 | 3.36 |
ENSRNOT00000090250
|
AABR07017783.4
|
|
| chr20_-_6960481 | 3.11 |
ENSRNOT00000093172
|
Mtch1
|
mitochondrial carrier 1 |
| chr1_+_69853553 | 2.95 |
ENSRNOT00000042086
|
Zfp954
|
zinc finger protein 954 |
| chr12_-_12407561 | 2.88 |
ENSRNOT00000037251
|
Bhlha15
|
basic helix-loop-helix family, member a15 |
| chr4_+_155437675 | 2.78 |
ENSRNOT00000031353
|
Dppa3
|
developmental pluripotency-associated 3 |
| chr3_-_79282493 | 2.55 |
ENSRNOT00000049832
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr2_-_59084059 | 2.49 |
ENSRNOT00000086323
ENSRNOT00000088701 |
Spef2
|
sperm flagellar 2 |
| chr1_+_167066516 | 2.43 |
ENSRNOT00000000474
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr18_+_55797198 | 2.42 |
ENSRNOT00000026334
ENSRNOT00000026394 |
Dctn4
|
dynactin subunit 4 |
| chr20_+_3148665 | 2.27 |
ENSRNOT00000086026
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr7_+_11054333 | 2.20 |
ENSRNOT00000007202
|
Gna15
|
guanine nucleotide binding protein, alpha 15 |
| chr2_-_59181863 | 1.89 |
ENSRNOT00000079636
|
Spef2
|
sperm flagellar 2 |
| chr1_+_213040024 | 1.77 |
ENSRNOT00000050237
|
Olr300
|
olfactory receptor 300 |
| chr7_+_142905758 | 1.70 |
ENSRNOT00000078663
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_+_209768696 | 1.68 |
ENSRNOT00000022406
|
Glrx3
|
glutaredoxin 3 |
| chr3_+_159887036 | 1.67 |
ENSRNOT00000031868
|
R3hdml
|
R3H domain containing-like |
| chr18_+_867048 | 1.64 |
ENSRNOT00000065494
|
Colec12
|
collectin sub-family member 12 |
| chr1_-_165606375 | 1.62 |
ENSRNOT00000024290
|
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr6_-_142534112 | 1.54 |
ENSRNOT00000073598
|
Ighv1-49
|
immunoglobulin heavy variable 1-49 |
| chr1_+_84612448 | 1.45 |
ENSRNOT00000032991
|
AABR07071891.1
|
|
| chr5_-_69517277 | 1.28 |
ENSRNOT00000079848
|
Olr850
|
olfactory receptor 850 |
| chr10_+_56453877 | 1.05 |
ENSRNOT00000031640
|
Plscr3
|
phospholipid scramblase 3 |
| chr9_+_20951260 | 0.85 |
ENSRNOT00000016906
|
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr19_-_37525762 | 0.85 |
ENSRNOT00000023606
|
Atp6v0d1
|
ATPase H+ transporting V0 subunit D1 |
| chr9_-_43127887 | 0.81 |
ENSRNOT00000021685
|
Ankrd39
|
ankyrin repeat domain 39 |
| chr5_+_162385173 | 0.80 |
ENSRNOT00000048232
|
Pramef12
|
PRAME family member 12 |
| chr5_+_167331035 | 0.51 |
ENSRNOT00000024443
|
Rere
|
arginine-glutamic acid dipeptide repeats |
| chr1_-_212912199 | 0.35 |
ENSRNOT00000045179
|
Olr295
|
olfactory receptor 295 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Smad3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.5 | 160.9 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 8.3 | 24.9 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 6.9 | 20.7 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 5.5 | 33.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 4.9 | 14.7 | GO:0015820 | leucine transport(GO:0015820) |
| 4.7 | 32.9 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 3.9 | 11.7 | GO:0036395 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) |
| 3.8 | 11.3 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 3.2 | 9.6 | GO:0032900 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) negative regulation of long term synaptic depression(GO:1900453) |
| 2.8 | 16.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 2.6 | 10.3 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 2.3 | 9.4 | GO:0038109 | Kit signaling pathway(GO:0038109) melanocyte migration(GO:0097324) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 2.0 | 15.9 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 1.8 | 32.7 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 1.8 | 174.1 | GO:0006342 | chromatin silencing(GO:0006342) |
| 1.4 | 11.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 1.3 | 12.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 1.2 | 12.8 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 1.0 | 85.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 1.0 | 16.0 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 1.0 | 9.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 1.0 | 10.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 1.0 | 7.7 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.9 | 2.8 | GO:0044726 | protection of DNA demethylation of female pronucleus(GO:0044726) |
| 0.8 | 4.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.7 | 20.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.6 | 29.0 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.6 | 38.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.5 | 18.5 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.5 | 6.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.5 | 6.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.5 | 8.4 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.4 | 12.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.3 | 7.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.3 | 1.7 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.3 | 1.6 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.3 | 4.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.3 | 2.9 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.3 | 1.7 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.3 | 4.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.3 | 2.6 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.2 | 2.4 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.2 | 20.2 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.2 | 5.7 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.2 | 27.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 2.5 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.2 | 0.5 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.2 | 2.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 3.7 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 2.4 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 5.9 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 1.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 4.7 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 3.9 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.1 | 4.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 17.3 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 2.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 3.1 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 15.5 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 2.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 4.3 | GO:0006397 | mRNA processing(GO:0006397) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 160.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 4.7 | 326.2 | GO:0000786 | nucleosome(GO:0000786) |
| 3.6 | 18.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 2.1 | 33.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 2.1 | 18.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.9 | 5.7 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 1.0 | 20.7 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.9 | 8.4 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.9 | 5.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.9 | 14.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.8 | 16.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.8 | 2.4 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.7 | 6.7 | GO:0031105 | septin complex(GO:0031105) |
| 0.6 | 9.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.5 | 9.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.4 | 7.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.4 | 4.4 | GO:0002177 | manchette(GO:0002177) |
| 0.4 | 11.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 2.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 12.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 9.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 6.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 2.8 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 34.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 16.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 4.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 4.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 32.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 28.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 10.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 38.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 15.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 2.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 12.8 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 7.7 | GO:0030425 | dendrite(GO:0030425) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 24.9 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 5.2 | 20.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 3.6 | 10.7 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 2.6 | 7.7 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 2.1 | 12.8 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.9 | 9.6 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.6 | 9.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.9 | 16.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.9 | 18.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.9 | 176.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.9 | 10.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.8 | 18.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.7 | 11.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.5 | 14.7 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.5 | 326.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.5 | 34.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.5 | 14.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 30.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.3 | 17.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.3 | 11.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 6.6 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.2 | 17.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.2 | 13.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.2 | 4.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 10.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 4.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 5.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 8.5 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 3.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 3.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 2.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 23.8 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 1.6 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) low-density lipoprotein particle binding(GO:0030169) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 3.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 15.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 2.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 12.1 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 8.4 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 4.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 18.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 6.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 2.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 6.3 | GO:0019904 | protein domain specific binding(GO:0019904) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 24.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.6 | 4.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.3 | 5.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 18.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 10.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 11.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 9.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 8.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 2.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 2.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 8.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 181.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 1.3 | 24.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 1.0 | 17.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.7 | 20.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.6 | 12.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.6 | 13.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.6 | 10.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.5 | 16.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 9.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.4 | 18.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 8.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 20.4 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.2 | 2.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.7 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 2.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |