Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
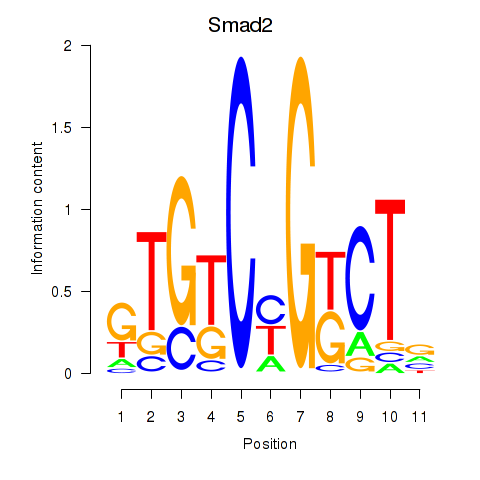
Results for Smad2
Z-value: 0.43
Transcription factors associated with Smad2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad2
|
ENSRNOG00000018140 | SMAD family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad2 | rn6_v1_chr18_+_72550219_72550219 | 0.03 | 6.5e-01 | Click! |
Activity profile of Smad2 motif
Sorted Z-values of Smad2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_253000760 | 22.97 |
ENSRNOT00000030024
|
Slc16a12
|
solute carrier family 16, member 12 |
| chr5_-_137372524 | 14.66 |
ENSRNOT00000009061
|
Tmem125
|
transmembrane protein 125 |
| chr4_-_117767772 | 12.87 |
ENSRNOT00000084170
|
LOC103690120
|
probable N-acetyltransferase CML1 |
| chr2_-_40386669 | 11.73 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr14_+_87312203 | 10.51 |
ENSRNOT00000088032
|
Adcy1
|
adenylate cyclase 1 |
| chr13_+_106463368 | 10.40 |
ENSRNOT00000003489
|
Esrrg
|
estrogen-related receptor gamma |
| chr2_-_220535751 | 7.43 |
ENSRNOT00000089082
|
Palmd
|
palmdelphin |
| chr4_-_117575154 | 6.94 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr1_+_100164400 | 6.84 |
ENSRNOT00000080334
|
LOC108348116
|
glandular kallikrein-3, submandibular-like |
| chr2_+_188449210 | 5.52 |
ENSRNOT00000027700
|
Pklr
|
pyruvate kinase, liver and RBC |
| chr17_+_81798756 | 4.97 |
ENSRNOT00000066826
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr18_-_77579969 | 4.57 |
ENSRNOT00000034896
|
Sall3
|
spalt-like transcription factor 3 |
| chr4_-_159192526 | 4.47 |
ENSRNOT00000026731
|
Kcna1
|
potassium voltage-gated channel subfamily A member 1 |
| chr4_+_113968995 | 4.20 |
ENSRNOT00000079511
|
Rtkn
|
rhotekin |
| chr9_+_17340341 | 4.15 |
ENSRNOT00000026637
ENSRNOT00000026559 ENSRNOT00000042790 ENSRNOT00000044163 ENSRNOT00000083811 |
Vegfa
|
vascular endothelial growth factor A |
| chr7_+_97559841 | 3.98 |
ENSRNOT00000007326
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr3_-_154042099 | 3.73 |
ENSRNOT00000033525
|
Blcap
|
bladder cancer associated protein |
| chr7_-_143523457 | 3.66 |
ENSRNOT00000012943
ENSRNOT00000082264 |
Krt4
|
keratin 4 |
| chr1_+_185427736 | 3.15 |
ENSRNOT00000064706
|
Plekha7
|
pleckstrin homology domain containing A7 |
| chr5_+_134737030 | 3.13 |
ENSRNOT00000066161
ENSRNOT00000057061 |
Kncn
|
kinocilin |
| chr15_+_12827707 | 2.80 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr6_+_27768943 | 2.71 |
ENSRNOT00000015820
|
Kif3c
|
kinesin family member 3C |
| chr14_+_84368476 | 2.56 |
ENSRNOT00000006827
|
Rnf215
|
ring finger protein 215 |
| chr7_-_70842405 | 2.37 |
ENSRNOT00000047449
|
Nxph4
|
neurexophilin 4 |
| chr2_-_149417212 | 1.99 |
ENSRNOT00000018573
|
Gpr87
|
G protein-coupled receptor 87 |
| chr3_+_172155496 | 1.21 |
ENSRNOT00000066279
|
Stx16
|
syntaxin 16 |
| chr13_-_91872954 | 1.18 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr18_+_30864216 | 1.06 |
ENSRNOT00000027015
|
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chr3_+_102723402 | 0.62 |
ENSRNOT00000040298
|
Olr765
|
olfactory receptor 765 |
| chr10_+_56453877 | 0.52 |
ENSRNOT00000031640
|
Plscr3
|
phospholipid scramblase 3 |
| chr1_-_170933660 | 0.43 |
ENSRNOT00000045360
|
Olr219
|
olfactory receptor 219 |
| chr7_-_139318455 | 0.31 |
ENSRNOT00000092029
|
Hdac7
|
histone deacetylase 7 |
| chr11_+_1896209 | 0.21 |
ENSRNOT00000000909
|
Cggbp1
|
CGG triplet repeat binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Smad2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.7 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.7 | 5.0 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 1.4 | 4.1 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) cardiac vascular smooth muscle cell development(GO:0060948) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.9 | 4.5 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.7 | 23.0 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.7 | 2.8 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.6 | 3.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.6 | 5.5 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.2 | 10.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 10.5 | GO:0007616 | long-term memory(GO:0007616) |
| 0.2 | 4.6 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.2 | 2.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 3.7 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.1 | 4.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 1.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 4.0 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 6.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 2.7 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.7 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 5.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 2.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.3 | 3.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.3 | 3.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 4.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 3.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 4.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 7.4 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 26.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 11.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 1.8 | 10.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 1.4 | 5.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.2 | 11.7 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.1 | 23.0 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.8 | 5.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.5 | 4.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.5 | 4.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.2 | 3.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 4.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 19.8 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.2 | 2.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 2.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 6.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 3.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 4.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 5.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.7 | 11.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 4.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 5.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 5.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 10.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 4.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |