Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
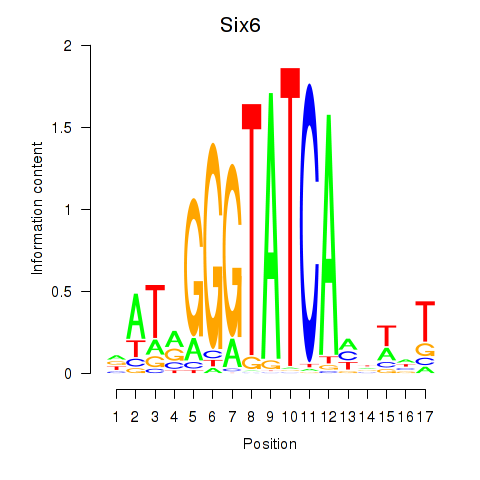
Results for Six6
Z-value: 0.87
Transcription factors associated with Six6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Six6
|
ENSRNOG00000006296 | SIX homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six6 | rn6_v1_chr6_+_95816749_95816749 | -0.16 | 5.3e-03 | Click! |
Activity profile of Six6 motif
Sorted Z-values of Six6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_75144903 | 45.92 |
ENSRNOT00000002329
|
Hrasls
|
HRAS-like suppressor |
| chr16_+_68633720 | 40.49 |
ENSRNOT00000081838
|
LOC100911229
|
sperm motility kinase-like |
| chr3_+_70327193 | 39.14 |
ENSRNOT00000089165
|
Fsip2
|
fibrous sheath-interacting protein 2 |
| chr11_+_52828116 | 38.57 |
ENSRNOT00000035340
|
Ccdc54
|
coiled-coil domain containing 54 |
| chr16_-_54628458 | 29.22 |
ENSRNOT00000042587
|
Adam24
|
ADAM metallopeptidase domain 24 |
| chr1_-_201702963 | 26.95 |
ENSRNOT00000031258
|
LOC499276
|
similar to RIKEN cDNA 1700022C21 |
| chr15_-_5417570 | 26.58 |
ENSRNOT00000061525
|
Spetex-2F
|
Spetex-2F protein |
| chr6_-_86223052 | 26.18 |
ENSRNOT00000046828
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr12_+_24158766 | 25.84 |
ENSRNOT00000001963
|
Ccl26
|
C-C motif chemokine ligand 26 |
| chr9_+_53440272 | 25.69 |
ENSRNOT00000059159
|
LOC685203
|
hypothetical protein LOC685203 |
| chr9_-_114327767 | 25.54 |
ENSRNOT00000085481
|
AABR07068674.1
|
|
| chr9_-_105376935 | 24.97 |
ENSRNOT00000043386
|
Slco6d1
|
solute carrier organic anion transporter family, member 6d1 |
| chr16_+_52169450 | 24.13 |
ENSRNOT00000019474
|
Triml1
|
tripartite motif family-like 1 |
| chr10_-_35058870 | 23.97 |
ENSRNOT00000079481
|
AABR07029573.3
|
|
| chr2_-_124047044 | 23.08 |
ENSRNOT00000065384
|
Cetn4
|
centrin 4 |
| chr4_+_45567573 | 20.89 |
ENSRNOT00000089824
|
Ankrd7
|
ankyrin repeat domain 7 |
| chr5_+_122508388 | 20.42 |
ENSRNOT00000038410
|
Tctex1d1
|
Tctex1 domain containing 1 |
| chr16_+_51730452 | 20.02 |
ENSRNOT00000078614
|
Adam34l
|
a disintegrin and metalloprotease domain 34-like |
| chr9_-_104870382 | 19.96 |
ENSRNOT00000084398
|
Slco6d1
|
solute carrier organic anion transporter family, member 6d1 |
| chr2_+_208541361 | 19.81 |
ENSRNOT00000021288
|
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chrX_+_147493880 | 19.61 |
ENSRNOT00000037881
|
MGC114427
|
similar to melanoma antigen family A, 5 |
| chr3_-_173944396 | 19.47 |
ENSRNOT00000079019
|
Sycp2
|
synaptonemal complex protein 2 |
| chr16_-_12194118 | 19.24 |
ENSRNOT00000071517
|
RGD1559804
|
similar to hypothetical protein 4930474N05 |
| chr11_-_1437732 | 19.10 |
ENSRNOT00000037345
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr1_+_106405002 | 19.10 |
ENSRNOT00000074027
|
LOC103690168
|
probable isocitrate dehydrogenase [NAD] gamma 2, mitochondrial |
| chr18_-_13194952 | 18.76 |
ENSRNOT00000092925
|
Ccdc178
|
coiled-coil domain containing 178 |
| chr5_+_162127810 | 18.75 |
ENSRNOT00000038858
|
Pramef27
|
PRAME family member 27 |
| chr2_+_92573004 | 18.48 |
ENSRNOT00000065774
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr3_+_161121697 | 18.03 |
ENSRNOT00000036020
|
Wfdc10a
|
WAP four-disulfide core domain 10A |
| chr2_-_137404996 | 17.91 |
ENSRNOT00000090907
|
LOC365791
|
similar to T-cell activation Rho GTPase-activating protein isoform b |
| chr9_+_17734345 | 17.89 |
ENSRNOT00000067104
|
Capn11
|
calpain 11 |
| chrY_+_380382 | 17.32 |
ENSRNOT00000092110
|
Rbmy1j
|
RNA binding motif protein, Y-linked, family 1, member J |
| chr2_+_127538659 | 17.27 |
ENSRNOT00000093483
ENSRNOT00000058476 |
Slc25a31
|
solute carrier family 25 member 31 |
| chr15_+_47470863 | 17.23 |
ENSRNOT00000072438
|
LOC683422
|
similar to Plasma kallikrein precursor (Plasma prekallikrein) (Kininogenin) (Fletcher factor) |
| chr11_-_36533073 | 17.00 |
ENSRNOT00000033486
|
Lca5l
|
LCA5L, lebercilin like |
| chr4_-_160803558 | 16.89 |
ENSRNOT00000049542
|
Senp18
|
Sumo1/sentrin/SMT3 specific peptidase 18 |
| chrX_+_125314613 | 16.84 |
ENSRNOT00000029193
|
LOC691215
|
hypothetical protein LOC691215 |
| chr13_-_100189778 | 16.66 |
ENSRNOT00000071062
|
LOC100912759
|
uncharacterized LOC100912759 |
| chr9_-_113246545 | 16.51 |
ENSRNOT00000034759
|
Tmem232
|
transmembrane protein 232 |
| chr10_-_91581232 | 16.32 |
ENSRNOT00000066453
ENSRNOT00000068686 |
AABR07030520.1
|
|
| chr18_+_47613854 | 16.28 |
ENSRNOT00000025270
|
Zfp474
|
zinc finger protein 474 |
| chr10_+_57239993 | 16.00 |
ENSRNOT00000067919
|
LOC687707
|
hypothetical protein LOC687707 |
| chr3_-_161040511 | 15.96 |
ENSRNOT00000037518
|
Wfdc6a
|
WAP four-disulfide core domain 6A |
| chr9_+_24066303 | 15.95 |
ENSRNOT00000018163
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chrX_+_43745604 | 15.91 |
ENSRNOT00000040825
|
LOC100362173
|
rCG36365-like |
| chr9_+_93445002 | 15.69 |
ENSRNOT00000025029
|
LOC501180
|
similar to hypothetical protein MGC35154 |
| chr4_-_161091010 | 15.43 |
ENSRNOT00000043708
|
Senp17
|
Sumo1/sentrin/SMT3 specific peptidase 17 |
| chr17_+_59714657 | 15.36 |
ENSRNOT00000035675
|
RGD1560860
|
similar to ankyrin repeat domain 26 |
| chr20_+_13498926 | 15.14 |
ENSRNOT00000070992
ENSRNOT00000045375 |
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr8_-_96547568 | 15.06 |
ENSRNOT00000078343
|
RGD1560775
|
similar to RIKEN cDNA 4930579C12 gene |
| chr5_+_69809495 | 14.98 |
ENSRNOT00000059828
|
Nipsnap3a
|
nipsnap homolog 3A (C. elegans) |
| chr6_-_55711146 | 14.94 |
ENSRNOT00000036996
|
Lrrc72
|
leucine rich repeat containing 72 |
| chr7_+_129595192 | 14.91 |
ENSRNOT00000071151
|
Zdhhc25
|
zinc finger, DHHC-type containing 25 |
| chr14_-_1505085 | 14.89 |
ENSRNOT00000090361
|
LOC102554799
|
uncharacterized LOC102554799 |
| chr5_-_58288125 | 14.82 |
ENSRNOT00000068752
|
Fam205a
|
family with sequence similarity 205, member A |
| chr14_+_107785029 | 14.78 |
ENSRNOT00000013097
ENSRNOT00000087091 |
Fam161a
|
family with sequence similarity 161, member A |
| chr15_+_33885106 | 14.73 |
ENSRNOT00000024483
|
Dhrs2
|
dehydrogenase/reductase 2 |
| chr17_+_5311274 | 14.56 |
ENSRNOT00000067020
|
LOC102547665
|
spermatogenesis-associated protein 31D1-like |
| chr11_-_88092756 | 14.16 |
ENSRNOT00000002542
|
Ydjc
|
YdjC homolog (bacterial) |
| chr18_-_13183263 | 13.88 |
ENSRNOT00000050933
|
Ccdc178
|
coiled-coil domain containing 178 |
| chr1_+_97712177 | 13.37 |
ENSRNOT00000027745
|
MGC114499
|
similar to RIKEN cDNA 4930433I11 gene |
| chrX_+_96667863 | 13.27 |
ENSRNOT00000042552
|
RGD1561151
|
similar to hypothetical protein 4932411N23 |
| chr2_-_148838441 | 13.14 |
ENSRNOT00000018294
|
Erich6
|
glutamate-rich 6 |
| chrX_+_78991223 | 13.06 |
ENSRNOT00000031200
|
Fam46d
|
family with sequence similarity 46, member D |
| chrX_+_151745455 | 12.89 |
ENSRNOT00000041429
|
LOC689396
|
similar to RNA polymerase II elongation factor ELL2 |
| chrX_+_74273182 | 12.88 |
ENSRNOT00000003922
|
Tsx
|
testis specific X-linked gene |
| chr3_-_14571640 | 12.87 |
ENSRNOT00000060013
|
Ggta1l1
|
glycoprotein, alpha-galactosyltransferase 1-like 1 |
| chr16_+_12174370 | 12.79 |
ENSRNOT00000072045
|
LOC102553538
|
ral guanine nucleotide dissociation stimulator-like |
| chr1_+_89149998 | 12.76 |
ENSRNOT00000056742
|
LOC688924
|
hypothetical protein LOC688924 |
| chr2_+_208749996 | 12.51 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr1_+_101412736 | 12.49 |
ENSRNOT00000067468
|
Lhb
|
luteinizing hormone beta polypeptide |
| chrX_-_38468360 | 12.43 |
ENSRNOT00000045285
|
Map7d2
|
MAP7 domain containing 2 |
| chr9_-_50820290 | 12.14 |
ENSRNOT00000050748
|
LOC102548013
|
uncharacterized LOC102548013 |
| chr15_+_17834635 | 11.99 |
ENSRNOT00000085530
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr6_-_105160470 | 11.97 |
ENSRNOT00000008367
|
Adam4
|
a disintegrin and metalloprotease domain 4 |
| chr5_-_107858104 | 11.87 |
ENSRNOT00000092196
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B |
| chr11_+_14005732 | 11.75 |
ENSRNOT00000047240
|
Rbm11
|
RNA binding motif protein 11 |
| chr14_-_77810147 | 11.59 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chr9_+_60883981 | 11.56 |
ENSRNOT00000081002
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr1_+_190914946 | 11.42 |
ENSRNOT00000039447
|
LOC691551
|
similar to F28B3.5a |
| chr11_-_61234944 | 11.41 |
ENSRNOT00000059680
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr14_+_83560541 | 11.23 |
ENSRNOT00000057738
ENSRNOT00000085228 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chrX_-_97262013 | 11.22 |
ENSRNOT00000040168
|
Cldn34c4
|
claudin 34C4 |
| chr10_+_90984227 | 11.08 |
ENSRNOT00000003890
ENSRNOT00000093707 |
Ccdc103
|
coiled-coil domain containing 103 |
| chr15_-_49666888 | 11.07 |
ENSRNOT00000041154
|
AABR07018269.1
|
|
| chr15_-_104168564 | 11.06 |
ENSRNOT00000093385
ENSRNOT00000038596 |
Dzip1
|
DAZ interacting zinc finger protein 1 |
| chr7_+_95309928 | 11.05 |
ENSRNOT00000005887
|
Mtbp
|
MDM2 binding protein |
| chr2_+_208750356 | 11.00 |
ENSRNOT00000041562
|
Chia
|
chitinase, acidic |
| chr1_-_87155118 | 10.95 |
ENSRNOT00000072441
|
AABR07002854.1
|
|
| chr7_-_130350570 | 10.82 |
ENSRNOT00000055805
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr13_-_47916185 | 10.82 |
ENSRNOT00000087793
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr2_+_189106039 | 10.80 |
ENSRNOT00000028210
|
Ube2q1
|
ubiquitin conjugating enzyme E2 Q1 |
| chr5_+_138245639 | 10.55 |
ENSRNOT00000066169
|
Svbp
|
small vasohibin binding protein |
| chr16_+_12510827 | 10.54 |
ENSRNOT00000077763
|
AABR07024716.1
|
|
| chrX_-_83788325 | 10.40 |
ENSRNOT00000037149
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr19_-_28148135 | 10.24 |
ENSRNOT00000090797
|
AABR07043407.1
|
|
| chr18_-_37776453 | 10.23 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr3_-_26056818 | 10.19 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chr1_-_90057923 | 10.07 |
ENSRNOT00000028674
|
Pdcd2l
|
programmed cell death 2-like |
| chr6_+_48452369 | 10.05 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chr13_-_6712734 | 9.99 |
ENSRNOT00000061683
|
AABR07019903.1
|
|
| chr9_+_88133884 | 9.88 |
ENSRNOT00000077265
|
Rhbdd1
|
rhomboid domain containing 1 |
| chr15_+_5783951 | 9.72 |
ENSRNOT00000061040
|
LOC305806
|
similar to glutaredoxin 1 (thioltransferase); glutaredoxin |
| chr11_-_90234286 | 9.60 |
ENSRNOT00000071986
|
Efcab1
|
EF hand calcium binding domain 1 |
| chr18_+_51523758 | 9.47 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr1_-_178195948 | 9.37 |
ENSRNOT00000081767
|
Btbd10
|
BTB domain containing 10 |
| chr4_-_80395502 | 9.26 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr1_-_100530183 | 9.26 |
ENSRNOT00000067754
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr16_+_13050305 | 9.25 |
ENSRNOT00000077388
|
AABR07024735.1
|
|
| chr17_-_42740021 | 9.24 |
ENSRNOT00000023063
|
Prl3a1
|
Prolactin family 3, subfamily a, member 1 |
| chr9_-_121725716 | 9.22 |
ENSRNOT00000087405
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr7_+_140248852 | 9.14 |
ENSRNOT00000087338
|
RGD1305928
|
hypothetical LOC300207 |
| chr8_-_16486419 | 9.07 |
ENSRNOT00000033223
|
LOC690235
|
similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase-like 1) |
| chr4_+_8066907 | 8.84 |
ENSRNOT00000080811
|
Srpk2
|
SRSF protein kinase 2 |
| chr14_-_21615488 | 8.82 |
ENSRNOT00000002667
|
Vcsa2
|
variable coding sequence A2 |
| chr18_-_67224566 | 8.77 |
ENSRNOT00000064947
|
Dcc
|
DCC netrin 1 receptor |
| chr1_+_102414625 | 8.75 |
ENSRNOT00000089488
|
Kcnc1
|
potassium voltage-gated channel subfamily C member 1 |
| chr6_-_1942972 | 8.54 |
ENSRNOT00000048711
|
Cdc42ep3
|
CDC42 effector protein 3 |
| chr1_-_99135977 | 8.54 |
ENSRNOT00000056515
|
Vom2r38
|
vomeronasal 2 receptor, 38 |
| chr18_+_84162467 | 8.49 |
ENSRNOT00000047841
|
RGD1559726
|
similar to melanoma antigen family A, 6 |
| chr19_+_26985450 | 8.47 |
ENSRNOT00000087468
|
AABR07043279.1
|
|
| chr2_-_137153551 | 8.42 |
ENSRNOT00000083443
|
AABR07010476.1
|
|
| chr5_+_124442293 | 8.38 |
ENSRNOT00000041922
|
RGD1564074
|
similar to novel protein |
| chr4_-_181348799 | 8.24 |
ENSRNOT00000033743
|
LOC690784
|
hypothetical protein LOC690783 |
| chr8_+_96395317 | 8.10 |
ENSRNOT00000044521
|
Trim43a
|
tripartite motif-containing 43A |
| chr20_-_13458110 | 8.08 |
ENSRNOT00000072183
|
Slc5a4
|
solute carrier family 5 member 4 |
| chr7_-_120874308 | 8.05 |
ENSRNOT00000078320
|
Fam227a
|
family with sequence similarity 227, member A |
| chr18_-_30802242 | 7.85 |
ENSRNOT00000064799
|
RGD1563159
|
RGD1563159 |
| chr8_+_91368111 | 7.81 |
ENSRNOT00000083151
|
Ttk
|
Ttk protein kinase |
| chr11_-_1836897 | 7.74 |
ENSRNOT00000050533
|
Zfp654
|
zinc finger protein 654 |
| chr8_-_37026142 | 7.65 |
ENSRNOT00000060446
|
LOC100361828
|
rCG22807-like |
| chr2_-_195279218 | 7.55 |
ENSRNOT00000085565
|
RGD1559714
|
similar to TDPOZ3 |
| chr14_-_78469480 | 7.55 |
ENSRNOT00000006771
|
Jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr1_+_215610368 | 7.49 |
ENSRNOT00000078903
ENSRNOT00000087781 |
Tnni2
|
troponin I2, fast skeletal type |
| chr9_+_23354770 | 7.45 |
ENSRNOT00000081459
|
Cenpq
|
centromere protein Q |
| chr13_+_71322759 | 7.41 |
ENSRNOT00000071815
|
Teddm1
|
transmembrane epididymal protein 1 |
| chr14_+_51959745 | 7.33 |
ENSRNOT00000083767
|
AABR07015320.1
|
|
| chr1_+_107262659 | 7.32 |
ENSRNOT00000022499
|
Gas2
|
growth arrest-specific 2 |
| chr2_+_211159062 | 7.22 |
ENSRNOT00000027099
|
Mybphl
|
myosin binding protein H-like |
| chr2_-_195935878 | 7.18 |
ENSRNOT00000028440
|
Cgn
|
cingulin |
| chr8_-_17524839 | 7.13 |
ENSRNOT00000081679
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr3_-_118635268 | 7.12 |
ENSRNOT00000086357
|
Atp8b4
|
ATPase phospholipid transporting 8B4 (putative) |
| chr8_-_111101416 | 7.09 |
ENSRNOT00000084413
|
Cep63
|
centrosomal protein 63 |
| chr10_+_53713938 | 7.01 |
ENSRNOT00000004236
ENSRNOT00000086599 ENSRNOT00000085582 |
Myh2
|
myosin heavy chain 2 |
| chr1_+_245237736 | 6.93 |
ENSRNOT00000035814
|
Vldlr
|
very low density lipoprotein receptor |
| chr8_-_5631014 | 6.88 |
ENSRNOT00000011884
|
Mmp1b
|
matrix metallopeptidase 1b (interstitial collagenase) |
| chr1_+_211351350 | 6.77 |
ENSRNOT00000082751
ENSRNOT00000030461 |
Jakmip3
|
janus kinase and microtubule interacting protein 3 |
| chr4_+_8066737 | 6.76 |
ENSRNOT00000063994
|
Srpk2
|
SRSF protein kinase 2 |
| chr14_-_37294472 | 6.68 |
ENSRNOT00000081087
|
Dcun1d4
|
defective in cullin neddylation 1 domain containing 4 |
| chr1_-_221087449 | 6.54 |
ENSRNOT00000016987
|
Fam89b
|
family with sequence similarity 89, member B |
| chr9_+_41045363 | 6.54 |
ENSRNOT00000066284
|
Cfc1
|
cripto, FRL-1, cryptic family 1 |
| chr10_+_16259728 | 6.33 |
ENSRNOT00000028115
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr8_-_5202360 | 6.26 |
ENSRNOT00000089124
|
AABR07069011.1
|
|
| chr2_-_185303610 | 6.18 |
ENSRNOT00000093479
ENSRNOT00000046286 |
NEWGENE_1592020
|
protease, serine 48 |
| chr5_-_74190991 | 6.14 |
ENSRNOT00000090366
|
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr9_+_95398237 | 6.10 |
ENSRNOT00000025879
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr15_+_18920373 | 6.06 |
ENSRNOT00000036754
|
LOC100363368
|
mCG11429-like |
| chr7_+_77678968 | 5.94 |
ENSRNOT00000006917
|
Atp6v1c1
|
ATPase H+ transporting V1 subunit C1 |
| chr9_-_19978013 | 5.88 |
ENSRNOT00000038222
|
Pla2g7
|
phospholipase A2 group VII |
| chr7_+_30699476 | 5.88 |
ENSRNOT00000059269
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr4_+_152630469 | 5.80 |
ENSRNOT00000013721
|
Ninj2
|
ninjurin 2 |
| chr20_+_14577166 | 5.72 |
ENSRNOT00000085583
|
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr19_+_15033108 | 5.67 |
ENSRNOT00000021812
|
Ces1d
|
carboxylesterase 1D |
| chr10_-_89338739 | 5.63 |
ENSRNOT00000073923
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr1_-_81127059 | 5.55 |
ENSRNOT00000026252
|
Zfp94
|
zinc finger protein 94 |
| chr12_-_17904254 | 5.54 |
ENSRNOT00000060192
|
LOC680273
|
similar to Forkhead box protein L1 (Forkhead-related protein FKHL11) (Forkhead-related transcription factor 7) (FREAC-7) |
| chr1_+_23409408 | 5.50 |
ENSRNOT00000022362
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr5_+_25391115 | 5.45 |
ENSRNOT00000021612
|
Cdh17
|
cadherin 17 |
| chr8_+_76426335 | 5.44 |
ENSRNOT00000085496
|
Gtf2a2
|
general transcription factor2A subunit 2 |
| chr20_-_795286 | 5.43 |
ENSRNOT00000052186
|
Olr1691
|
olfactory receptor 1691 |
| chrX_+_145558840 | 5.37 |
ENSRNOT00000032522
|
4931400O07Rik
|
RIKEN cDNA 4931400O07 gene |
| chrX_-_77529374 | 5.34 |
ENSRNOT00000078213
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr19_-_49623758 | 5.29 |
ENSRNOT00000071130
|
Pkd1l2
|
polycystic kidney disease 1-like 2 |
| chr10_-_16689321 | 5.26 |
ENSRNOT00000028173
|
Bnip1
|
BCL2/adenovirus E1B interacting protein 1 |
| chr10_-_13107771 | 5.23 |
ENSRNOT00000005879
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr2_+_150106292 | 5.19 |
ENSRNOT00000073078
|
LOC100910567
|
arylacetamide deacetylase-like 2-like |
| chr10_+_103972888 | 5.19 |
ENSRNOT00000067838
|
Kctd2
|
potassium channel tetramerization domain containing 2 |
| chr17_+_35677984 | 5.14 |
ENSRNOT00000024609
|
Uqcrfs1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr1_+_171188133 | 5.08 |
ENSRNOT00000026511
|
Olr231
|
olfactory receptor 231 |
| chr5_-_77945016 | 5.07 |
ENSRNOT00000076223
|
Zfp37
|
zinc finger protein 37 |
| chr17_-_42737000 | 4.98 |
ENSRNOT00000077923
|
Prl3a1
|
Prolactin family 3, subfamily a, member 1 |
| chr19_+_15195565 | 4.87 |
ENSRNOT00000090865
ENSRNOT00000078874 |
Ces1d
|
carboxylesterase 1D |
| chr5_-_169301728 | 4.83 |
ENSRNOT00000013646
|
Espn
|
espin |
| chr8_+_84945444 | 4.82 |
ENSRNOT00000008244
|
Klhl31
|
kelch-like family member 31 |
| chr3_-_102484517 | 4.81 |
ENSRNOT00000006522
|
Olr750
|
olfactory receptor 750 |
| chrX_-_77418525 | 4.78 |
ENSRNOT00000092303
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr4_-_67206426 | 4.77 |
ENSRNOT00000013124
|
Mkrn1
|
makorin ring finger protein 1 |
| chr2_+_5437067 | 4.63 |
ENSRNOT00000093516
|
Pou5f2
|
POU domain class 5, transcription factor 2 |
| chr3_+_56966654 | 4.48 |
ENSRNOT00000088097
|
Gorasp2
|
golgi reassembly stacking protein 2 |
| chr5_-_157059508 | 4.42 |
ENSRNOT00000022282
|
Vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chrX_-_142248369 | 4.33 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr7_-_117773134 | 4.24 |
ENSRNOT00000045135
|
Recql4
|
RecQ like helicase 4 |
| chr10_+_106991935 | 4.15 |
ENSRNOT00000004007
|
Pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr1_+_27476375 | 4.14 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr8_-_71055969 | 4.12 |
ENSRNOT00000075734
|
Ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr17_+_77195247 | 4.06 |
ENSRNOT00000081940
|
Optn
|
optineurin |
| chr3_-_94419048 | 4.06 |
ENSRNOT00000015775
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr11_-_61470348 | 4.05 |
ENSRNOT00000083841
|
LOC102553099
|
N-alpha-acetyltransferase 50-like |
| chr16_-_19544727 | 4.01 |
ENSRNOT00000074514
|
Zfp617
|
zinc finger protein 617 |
| chr7_-_95310005 | 3.98 |
ENSRNOT00000005815
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Six6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 29.2 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 4.7 | 23.5 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 3.8 | 45.9 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 3.7 | 11.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 3.5 | 10.5 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 3.3 | 9.9 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 3.1 | 15.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 2.8 | 16.9 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 2.6 | 7.8 | GO:0044785 | metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 2.5 | 49.6 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 2.3 | 9.2 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 2.2 | 8.8 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 2.2 | 6.5 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 2.2 | 10.8 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 2.1 | 10.4 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 1.9 | 11.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 1.8 | 5.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 1.7 | 6.9 | GO:0034436 | glycoprotein transport(GO:0034436) reelin-mediated signaling pathway(GO:0038026) |
| 1.6 | 16.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.6 | 6.5 | GO:0060459 | left lung development(GO:0060459) left lung morphogenesis(GO:0060460) |
| 1.6 | 3.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 1.5 | 5.9 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) platelet activating factor metabolic process(GO:0046469) |
| 1.5 | 8.7 | GO:0021759 | globus pallidus development(GO:0021759) |
| 1.4 | 25.8 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 1.2 | 6.1 | GO:0050955 | thermoception(GO:0050955) |
| 1.2 | 10.8 | GO:0070459 | prolactin secretion(GO:0070459) |
| 1.2 | 12.9 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 1.2 | 7.0 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 1.1 | 11.1 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 1.0 | 7.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.9 | 9.3 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.9 | 10.5 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.9 | 2.6 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.8 | 2.4 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.7 | 4.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.7 | 4.8 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.7 | 10.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.7 | 3.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.7 | 3.4 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.7 | 2.6 | GO:0061055 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) myotome development(GO:0061055) |
| 0.6 | 17.9 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.6 | 3.1 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.6 | 4.3 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.6 | 6.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.6 | 5.5 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.6 | 1.8 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.6 | 3.0 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.6 | 6.9 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.6 | 3.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.5 | 11.4 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.5 | 4.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.5 | 1.5 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.5 | 1.9 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.5 | 2.8 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.5 | 11.9 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.4 | 13.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.4 | 4.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.4 | 1.6 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.4 | 2.4 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.4 | 12.1 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.4 | 13.5 | GO:0030823 | regulation of cGMP metabolic process(GO:0030823) |
| 0.4 | 1.5 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 2.7 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.3 | 3.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 6.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.3 | 11.1 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.3 | 4.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.3 | 3.6 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.3 | 3.4 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.3 | 3.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.0 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.2 | 5.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.2 | 3.9 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.2 | 2.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 1.0 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 3.3 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 1.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 3.6 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.2 | 0.7 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.2 | 0.5 | GO:0060545 | positive regulation of necroptotic process(GO:0060545) |
| 0.2 | 1.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 1.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.6 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 7.2 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.9 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 3.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 1.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 6.3 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 3.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 3.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.5 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 1.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 5.3 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 2.7 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 0.5 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 5.8 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.1 | 14.9 | GO:0044782 | cilium organization(GO:0044782) |
| 0.1 | 4.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 14.1 | GO:0009566 | fertilization(GO:0009566) |
| 0.1 | 5.9 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 6.2 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 3.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.8 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.9 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.1 | 0.8 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 2.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.9 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 1.0 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.9 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 1.2 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.1 | 6.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 4.5 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 3.8 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.3 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 1.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 1.0 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 1.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 3.0 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.8 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 10.1 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 4.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 8.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 3.2 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 0.3 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 15.7 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 0.8 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 45.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 2.8 | 49.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 1.5 | 26.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 1.4 | 16.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.2 | 9.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 1.2 | 5.9 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 1.1 | 8.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.9 | 15.9 | GO:0042581 | specific granule(GO:0042581) |
| 0.9 | 19.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.9 | 11.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.8 | 7.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.6 | 7.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.5 | 3.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.5 | 5.9 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.5 | 5.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.4 | 3.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.4 | 4.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.4 | 16.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.4 | 1.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.4 | 4.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.4 | 17.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 3.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 2.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 2.0 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.3 | 47.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.3 | 3.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.3 | 2.8 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.3 | 37.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 10.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 8.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 9.8 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 2.8 | GO:0051286 | cell tip(GO:0051286) |
| 0.2 | 28.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.2 | 3.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 11.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 1.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 1.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 2.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 2.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 2.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 0.5 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 10.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 8.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 8.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 10.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 7.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 6.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 5.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 4.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 5.1 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 8.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 1.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 15.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 3.4 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 0.5 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 2.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 2.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 4.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 4.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 5.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 3.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 26.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 25.8 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 4.8 | 19.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 3.9 | 23.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 3.5 | 10.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 3.3 | 49.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 3.2 | 12.9 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 2.9 | 14.7 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 2.6 | 10.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 2.3 | 6.9 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 1.8 | 5.4 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 1.8 | 8.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 1.4 | 13.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 1.3 | 10.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.3 | 16.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.1 | 7.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 1.1 | 8.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.0 | 25.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 1.0 | 5.9 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.9 | 2.6 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.9 | 7.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.8 | 11.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.8 | 6.5 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.8 | 4.1 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 0.8 | 2.4 | GO:0099567 | N-terminal myristoylation domain binding(GO:0031997) calcium ion binding involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099567) |
| 0.7 | 17.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.7 | 4.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.7 | 11.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.7 | 2.7 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.7 | 5.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.6 | 5.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.6 | 7.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.6 | 11.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.6 | 2.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.5 | 3.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.5 | 65.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.5 | 4.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.5 | 3.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.5 | 1.5 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.5 | 13.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.4 | 21.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.4 | 8.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.4 | 8.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.4 | 13.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.4 | 3.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 9.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.3 | 3.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 32.1 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.3 | 10.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 6.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 2.8 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.3 | 10.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.3 | 11.2 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.3 | 3.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 1.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 3.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 5.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 1.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 24.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.2 | 3.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.2 | 5.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 4.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 3.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 5.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 3.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 2.9 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 23.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 15.3 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.1 | 0.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 3.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 0.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 3.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 11.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) TBP-class protein binding(GO:0017025) |
| 0.0 | 8.3 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 23.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.0 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 5.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.6 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 1.8 | GO:0008233 | peptidase activity(GO:0008233) |
| 0.0 | 6.8 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 25.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.3 | 11.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 8.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 7.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 3.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 7.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 7.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.8 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 8.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 12.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.7 | 9.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.4 | 16.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 5.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.4 | 7.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 9.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 19.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 3.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.3 | 7.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.3 | 7.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 3.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 2.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 9.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 3.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 3.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 8.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 7.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 3.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 9.1 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.1 | 2.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 2.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 5.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |