Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
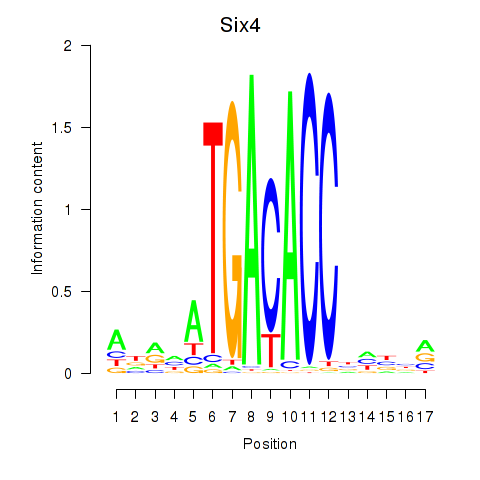
Results for Six4
Z-value: 0.78
Transcription factors associated with Six4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Six4
|
ENSRNOG00000007250 | SIX homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six4 | rn6_v1_chr6_-_95998529_95998529 | -0.02 | 7.3e-01 | Click! |
Activity profile of Six4 motif
Sorted Z-values of Six4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_39958239 | 24.97 |
ENSRNOT00000050368
|
Myl2
|
myosin light chain 2 |
| chr1_+_12915734 | 22.65 |
ENSRNOT00000089066
|
Txlnb
|
taxilin beta |
| chr6_-_139041812 | 19.00 |
ENSRNOT00000074510
|
AABR07065656.1
|
|
| chr16_-_79671719 | 18.78 |
ENSRNOT00000015908
|
Myom2
|
myomesin 2 |
| chr6_-_139041654 | 17.72 |
ENSRNOT00000075664
|
AABR07065656.1
|
|
| chr6_-_139102378 | 17.48 |
ENSRNOT00000086423
|
AABR07065656.5
|
|
| chr8_-_111965889 | 17.36 |
ENSRNOT00000032376
|
Bfsp2
|
beaded filament structural protein 2 |
| chr6_-_141147264 | 16.88 |
ENSRNOT00000042900
|
LOC100361105
|
Igh protein-like |
| chr6_-_138744480 | 15.19 |
ENSRNOT00000089387
|
AABR07065651.5
|
|
| chr3_+_19274273 | 15.13 |
ENSRNOT00000040102
|
AABR07051684.1
|
|
| chr6_-_138852571 | 14.11 |
ENSRNOT00000081803
|
AABR07065656.8
|
|
| chr6_-_141321108 | 13.56 |
ENSRNOT00000040556
|
AABR07065789.3
|
|
| chr6_-_41870046 | 12.44 |
ENSRNOT00000005863
|
Lpin1
|
lipin 1 |
| chr7_-_144880092 | 10.66 |
ENSRNOT00000055281
|
Nfe2
|
nuclear factor, erythroid 2 |
| chr1_-_141893674 | 10.50 |
ENSRNOT00000019059
ENSRNOT00000085988 |
Idh2
|
isocitrate dehydrogenase (NADP(+)) 2, mitochondrial |
| chr1_-_209641123 | 9.93 |
ENSRNOT00000021702
|
Ebf3
|
early B-cell factor 3 |
| chr8_+_130538651 | 9.68 |
ENSRNOT00000026343
|
Ackr2
|
atypical chemokine receptor 2 |
| chr10_-_14703668 | 8.64 |
ENSRNOT00000024767
|
Tpsab1
|
tryptase alpha/beta 1 |
| chr13_-_47979797 | 8.58 |
ENSRNOT00000080035
|
Rassf5
|
Ras association domain family member 5 |
| chr6_-_141198565 | 8.10 |
ENSRNOT00000064361
|
AABR07065782.1
|
|
| chr12_-_50400765 | 7.71 |
ENSRNOT00000071613
|
Crybb1
|
crystallin, beta B1 |
| chr1_-_221087449 | 7.24 |
ENSRNOT00000016987
|
Fam89b
|
family with sequence similarity 89, member B |
| chrX_-_72132495 | 7.09 |
ENSRNOT00000076391
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr10_-_107114271 | 6.98 |
ENSRNOT00000004035
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr1_+_221773254 | 6.96 |
ENSRNOT00000028646
|
Rasgrp2
|
RAS guanyl releasing protein 2 |
| chr11_-_61530567 | 6.74 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr2_+_127845034 | 6.50 |
ENSRNOT00000044804
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr14_-_21709084 | 6.45 |
ENSRNOT00000087477
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chr15_+_5319916 | 6.44 |
ENSRNOT00000046644
|
LOC102546495
|
disks large homolog 5-like |
| chr7_+_120580743 | 6.17 |
ENSRNOT00000017181
|
Maff
|
MAF bZIP transcription factor F |
| chr5_-_152247332 | 5.95 |
ENSRNOT00000077165
|
Lin28a
|
lin-28 homolog A |
| chr17_-_15566332 | 5.91 |
ENSRNOT00000093743
|
Ecm2
|
extracellular matrix protein 2 |
| chr1_-_89045586 | 5.80 |
ENSRNOT00000063808
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr9_+_60883981 | 5.66 |
ENSRNOT00000081002
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr1_-_53038229 | 5.44 |
ENSRNOT00000017282
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr6_+_110749705 | 5.18 |
ENSRNOT00000084348
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr10_-_104624757 | 5.04 |
ENSRNOT00000087759
|
Unc13d
|
unc-13 homolog D |
| chr3_-_94418711 | 4.97 |
ENSRNOT00000089554
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr9_+_17728816 | 4.84 |
ENSRNOT00000065754
|
Capn11
|
calpain 11 |
| chr1_-_166037424 | 4.81 |
ENSRNOT00000026115
|
P2ry2
|
purinergic receptor P2Y2 |
| chr7_+_72599479 | 4.75 |
ENSRNOT00000018963
|
AABR07057460.1
|
|
| chrX_+_583925 | 4.58 |
ENSRNOT00000075665
|
AABR07036698.1
|
|
| chr15_+_24254042 | 4.50 |
ENSRNOT00000092161
|
Fbxo34
|
F-box protein 34 |
| chr13_+_92103123 | 4.49 |
ENSRNOT00000004643
|
Olr1584
|
olfactory receptor 1584 |
| chr15_+_24267323 | 4.37 |
ENSRNOT00000015778
|
Fbxo34
|
F-box protein 34 |
| chr4_-_153465203 | 4.26 |
ENSRNOT00000016776
|
Bid
|
BH3 interacting domain death agonist |
| chr1_+_141218095 | 4.24 |
ENSRNOT00000051411
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr1_-_229601032 | 4.23 |
ENSRNOT00000016690
|
Cntf
|
ciliary neurotrophic factor |
| chr4_-_98593664 | 4.22 |
ENSRNOT00000007927
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr10_-_40381886 | 4.16 |
ENSRNOT00000050213
|
LOC100912427
|
nuclease-sensitive element-binding protein 1-like |
| chr3_-_94419048 | 3.76 |
ENSRNOT00000015775
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr6_-_141384305 | 3.75 |
ENSRNOT00000045320
|
AABR07065790.1
|
|
| chr7_+_11660934 | 3.69 |
ENSRNOT00000022336
|
Lmnb2
|
lamin B2 |
| chr3_+_117853999 | 3.55 |
ENSRNOT00000042949
|
LOC100911847
|
40S ribosomal protein S14-like |
| chr3_-_153321352 | 3.54 |
ENSRNOT00000064824
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr3_+_111826297 | 3.52 |
ENSRNOT00000081897
|
Pla2g4b
|
phospholipase A2 group IVB |
| chr16_-_81434363 | 3.44 |
ENSRNOT00000080963
|
Rasa3
|
RAS p21 protein activator 3 |
| chr8_+_32165810 | 3.40 |
ENSRNOT00000007539
|
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr1_-_190596431 | 3.34 |
ENSRNOT00000079335
|
Pdzd9
|
PDZ domain containing 9 |
| chr13_-_73921969 | 3.32 |
ENSRNOT00000090848
|
Tdrd5
|
tudor domain containing 5 |
| chr1_-_101175570 | 3.09 |
ENSRNOT00000073918
|
Gfy
|
golgi-associated, olfactory signaling regulator |
| chr10_+_103545439 | 2.78 |
ENSRNOT00000066523
|
Cd300ld
|
Cd300 molecule-like family member D |
| chrX_-_123018444 | 2.73 |
ENSRNOT00000071692
|
Gzmbl1
|
Granszyme B-like 1 |
| chr14_+_77079402 | 2.68 |
ENSRNOT00000042200
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr1_-_73121085 | 2.54 |
ENSRNOT00000073464
|
LOC684545
|
similar to NACHT, leucine rich repeat and PYD containing 2 |
| chr3_-_147985616 | 2.50 |
ENSRNOT00000087431
|
Defb26
|
defensin beta 26 |
| chrX_-_121066855 | 2.37 |
ENSRNOT00000044165
|
Tesl
|
testis derived transcript-like |
| chr1_-_99135977 | 2.25 |
ENSRNOT00000056515
|
Vom2r38
|
vomeronasal 2 receptor, 38 |
| chr14_-_43890171 | 2.22 |
ENSRNOT00000003382
|
Chrna9
|
cholinergic receptor nicotinic alpha 9 subunit |
| chr3_-_158985814 | 2.20 |
ENSRNOT00000071070
|
LOC100909423
|
olfactory receptor 150-like |
| chr5_+_138354324 | 2.12 |
ENSRNOT00000066742
|
Gm17728
|
predicted gene, 17728 |
| chr2_-_194084716 | 1.78 |
ENSRNOT00000049525
|
AABR07012342.1
|
|
| chr14_+_2613406 | 1.67 |
ENSRNOT00000000083
|
Tmed5
|
transmembrane p24 trafficking protein 5 |
| chr4_+_143911791 | 1.62 |
ENSRNOT00000081511
|
AABR07061780.1
|
|
| chr17_+_72240802 | 1.60 |
ENSRNOT00000025993
ENSRNOT00000090453 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr13_-_73921303 | 1.60 |
ENSRNOT00000005353
|
Tdrd5
|
tudor domain containing 5 |
| chr1_-_71909138 | 1.54 |
ENSRNOT00000029591
|
Nlrp9
|
NLR family, pyrin domain containing 9 |
| chr1_+_248195797 | 1.52 |
ENSRNOT00000066891
|
LOC103690131
|
tumor protein D55-like |
| chr7_-_69982592 | 1.50 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr17_-_42926523 | 1.45 |
ENSRNOT00000022512
|
Prl3d4
|
prolactin family 3, subfamily d, member 4 |
| chr10_+_4447275 | 1.43 |
ENSRNOT00000087196
ENSRNOT00000074890 |
Rsl1d1l1
|
ribosomal L1 domain containing 1-like 1 |
| chr10_-_86645529 | 1.37 |
ENSRNOT00000011947
|
Med24
|
mediator complex subunit 24 |
| chr3_-_93734282 | 1.34 |
ENSRNOT00000012428
|
Caprin1
|
cell cycle associated protein 1 |
| chr10_-_12214822 | 1.30 |
ENSRNOT00000071005
|
Olr1365
|
olfactory receptor 1365 |
| chr1_-_32272476 | 1.28 |
ENSRNOT00000022683
|
Tert
|
telomerase reverse transcriptase |
| chr8_+_20358272 | 1.25 |
ENSRNOT00000071855
|
Olr1169
|
olfactory receptor 1169 |
| chr5_+_159671136 | 0.96 |
ENSRNOT00000084737
|
Fbxo42
|
F-box protein 42 |
| chr17_-_42740021 | 0.95 |
ENSRNOT00000023063
|
Prl3a1
|
Prolactin family 3, subfamily a, member 1 |
| chr1_+_80135391 | 0.92 |
ENSRNOT00000021893
|
Gpr4
|
G protein-coupled receptor 4 |
| chr1_+_70260041 | 0.81 |
ENSRNOT00000020416
|
Zim1
|
zinc finger, imprinted 1 |
| chr10_+_13289508 | 0.79 |
ENSRNOT00000085694
|
Sbp
|
spermine binding protein |
| chr5_-_157368450 | 0.67 |
ENSRNOT00000023143
ENSRNOT00000079855 |
Otud3
|
OTU deubiquitinase 3 |
| chr8_-_77398156 | 0.51 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr20_+_5040337 | 0.36 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr1_+_79396578 | 0.27 |
ENSRNOT00000023686
|
Ceacam11
|
carcinoembryonic antigen-related cell adhesion molecule 11 |
| chr1_+_99616447 | 0.25 |
ENSRNOT00000029197
|
Klk14
|
kallikrein related-peptidase 14 |
| chr14_+_113530470 | 0.21 |
ENSRNOT00000004919
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr17_-_42817037 | 0.05 |
ENSRNOT00000023166
|
Prl3c1
|
Prolactin family 3, subfamily c, member 1 |
| chr1_-_167971151 | 0.02 |
ENSRNOT00000043023
|
Olr53
|
olfactory receptor 53 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Six4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 25.0 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 2.7 | 18.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 2.6 | 10.5 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 2.4 | 7.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.8 | 5.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 1.8 | 12.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 1.7 | 5.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 1.6 | 4.9 | GO:0007308 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 1.4 | 7.1 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) response to interleukin-9(GO:0071104) |
| 1.2 | 17.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 1.1 | 10.7 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 1.1 | 4.2 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 1.1 | 4.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.9 | 6.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.8 | 6.7 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.7 | 4.8 | GO:0071415 | cellular response to purine-containing compound(GO:0071415) |
| 0.6 | 4.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.6 | 8.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.5 | 4.3 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.4 | 1.3 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.2 | 0.7 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.2 | 4.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.2 | 0.9 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.2 | 3.1 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.1 | 2.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 9.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 2.5 | GO:0050718 | positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 | 1.4 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.1 | 6.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 3.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 6.5 | GO:0051931 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 7.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 2.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 1.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 8.6 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 1.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 3.4 | GO:0046580 | cellular response to heat(GO:0034605) negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 1.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 6.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 5.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 2.8 | GO:0032680 | regulation of tumor necrosis factor production(GO:0032680) |
| 0.0 | 1.4 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 5.8 | GO:0042098 | T cell proliferation(GO:0042098) |
| 0.0 | 0.3 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 3.9 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 2.5 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 2.7 | GO:0016485 | protein processing(GO:0016485) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 25.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.7 | 18.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.4 | 4.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.0 | 6.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.8 | 5.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.5 | 4.9 | GO:0071546 | pi-body(GO:0071546) |
| 0.5 | 4.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.5 | 3.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.3 | 8.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 5.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 3.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 5.8 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.2 | 5.4 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 18.1 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 7.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 4.3 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 17.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 12.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 1.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 8.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.2 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.1 | 7.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 3.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 10.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 4.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.5 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 4.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 5.8 | GO:0000228 | nuclear chromosome(GO:0000228) |
| 0.0 | 3.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 3.1 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 10.5 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 1.6 | 4.8 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 1.4 | 18.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.2 | 5.9 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.1 | 25.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.1 | 5.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.0 | 9.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.8 | 25.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.7 | 6.0 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.7 | 12.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.7 | 7.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.7 | 4.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.5 | 6.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 4.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.4 | 3.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.4 | 7.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 10.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 2.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 1.4 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.2 | 4.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 3.4 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.2 | 1.3 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 2.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 3.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 15.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 3.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 1.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.8 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 3.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 17.8 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 7.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 5.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 7.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 6.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 9.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 8.7 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 4.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 25.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 4.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 15.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 10.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 3.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 5.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 4.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 12.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.6 | 25.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 10.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.6 | 5.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 7.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.4 | 4.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.3 | 8.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 4.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 2.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 3.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 16.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 4.2 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 1.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |