Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Six3_Six1_Six2
Z-value: 0.60
Transcription factors associated with Six3_Six1_Six2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
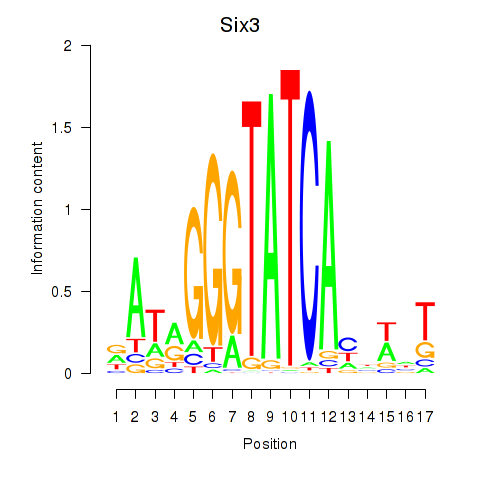
Six3
|
ENSRNOG00000057031 | SIX homeobox 3 |
|
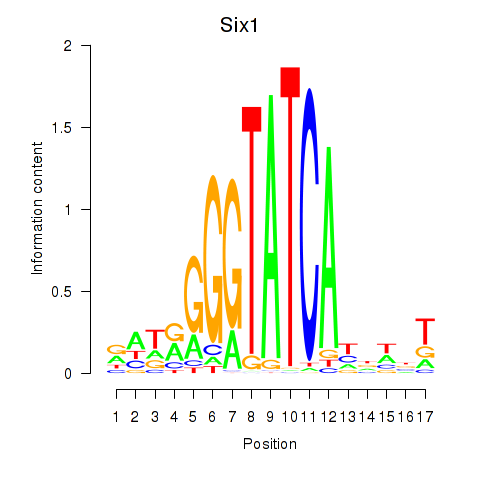
Six1
|
ENSRNOG00000022777 | SIX homeobox 1 |
|
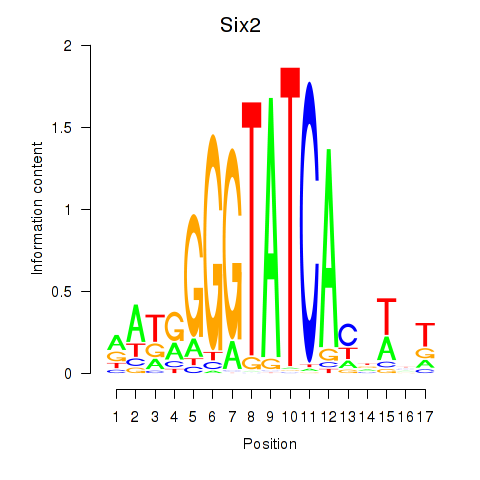
Six2
|
ENSRNOG00000058827 | SIX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six3 | rn6_v1_chr6_+_8886591_8886730 | -0.41 | 1.6e-14 | Click! |
| Six1 | rn6_v1_chr6_-_95934296_95934296 | 0.11 | 6.0e-02 | Click! |
| Six2 | rn6_v1_chr6_-_8956276_8956276 | -0.08 | 1.3e-01 | Click! |
Activity profile of Six3_Six1_Six2 motif
Sorted Z-values of Six3_Six1_Six2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_141893674 | 39.43 |
ENSRNOT00000019059
ENSRNOT00000085988 |
Idh2
|
isocitrate dehydrogenase (NADP(+)) 2, mitochondrial |
| chr8_+_33514042 | 37.26 |
ENSRNOT00000081614
ENSRNOT00000081525 |
Kcnj1
|
potassium voltage-gated channel subfamily J member 1 |
| chr4_-_51199570 | 28.67 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr2_-_53413638 | 26.88 |
ENSRNOT00000021081
|
Ghr
|
growth hormone receptor |
| chr1_-_198104109 | 22.55 |
ENSRNOT00000026186
|
Sult1a1
|
sulfotransferase family 1A member 1 |
| chr1_-_189182306 | 21.56 |
ENSRNOT00000021249
|
Gp2
|
glycoprotein 2 |
| chr5_+_118743632 | 21.21 |
ENSRNOT00000013785
|
Pgm1
|
phosphoglucomutase 1 |
| chr1_-_189181901 | 18.39 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr10_+_53713938 | 17.18 |
ENSRNOT00000004236
ENSRNOT00000086599 ENSRNOT00000085582 |
Myh2
|
myosin heavy chain 2 |
| chr17_-_46115004 | 16.78 |
ENSRNOT00000087838
|
Aoah
|
acyloxyacyl hydrolase |
| chr4_-_155867708 | 16.34 |
ENSRNOT00000051525
|
Clec4a2
|
C-type lectin domain family 4, member A2 |
| chr18_-_40716686 | 15.08 |
ENSRNOT00000000172
|
Cdo1
|
cysteine dioxygenase type 1 |
| chr15_+_57290849 | 14.44 |
ENSRNOT00000014909
|
Cpb2
|
carboxypeptidase B2 |
| chr13_-_95943761 | 13.19 |
ENSRNOT00000005961
|
Adss
|
adenylosuccinate synthase |
| chrX_+_156336450 | 12.28 |
ENSRNOT00000082124
|
Slc10a3
|
solute carrier family 10, member 3 |
| chrX_-_114232939 | 12.21 |
ENSRNOT00000042639
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr5_+_158090173 | 10.89 |
ENSRNOT00000088766
ENSRNOT00000079516 ENSRNOT00000092026 |
Tas1r2
|
taste 1 receptor member 2 |
| chr4_+_156050054 | 9.74 |
ENSRNOT00000039855
|
Clec4a
|
C-type lectin domain family 4, member A |
| chr1_-_13341952 | 9.51 |
ENSRNOT00000079531
|
NEWGENE_2319083
|
epithelial cell transforming 2 like |
| chrX_+_16050780 | 9.23 |
ENSRNOT00000079054
|
Clcn5
|
chloride voltage-gated channel 5 |
| chr1_-_76517134 | 8.28 |
ENSRNOT00000064593
ENSRNOT00000085775 |
LOC100912485
|
alcohol sulfotransferase-like |
| chr10_+_70298519 | 7.93 |
ENSRNOT00000035730
ENSRNOT00000076233 ENSRNOT00000076445 ENSRNOT00000076133 |
Slfn5
|
schlafen family member 5 |
| chr17_-_32076181 | 7.86 |
ENSRNOT00000074842
|
LOC100911107
|
leukocyte elastase inhibitor A-like |
| chr1_-_167700332 | 7.31 |
ENSRNOT00000092890
|
Trim21
|
tripartite motif-containing 21 |
| chr2_+_44664124 | 7.19 |
ENSRNOT00000066098
|
Plpp1
|
phospholipid phosphatase 1 |
| chr1_+_201981357 | 6.90 |
ENSRNOT00000027999
|
Acadsb
|
acyl-CoA dehydrogenase, short/branched chain |
| chr2_-_182846061 | 6.67 |
ENSRNOT00000013025
|
Tlr2
|
toll-like receptor 2 |
| chr11_-_61470046 | 6.49 |
ENSRNOT00000073436
|
LOC102553099
|
N-alpha-acetyltransferase 50-like |
| chr1_-_213650247 | 6.28 |
ENSRNOT00000019679
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb |
| chr1_-_100530183 | 6.06 |
ENSRNOT00000067754
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr5_+_124442293 | 5.96 |
ENSRNOT00000041922
|
RGD1564074
|
similar to novel protein |
| chr8_+_70630767 | 5.88 |
ENSRNOT00000051353
|
Igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chrX_+_123912361 | 5.77 |
ENSRNOT00000092622
|
Rhox5
|
reproductive homeobox 5 |
| chr4_+_1436254 | 5.72 |
ENSRNOT00000088564
|
Olr1232
|
olfactory receptor 1232 |
| chr9_-_92524739 | 5.68 |
ENSRNOT00000089889
|
Slc16a14
|
solute carrier family 16, member 14 |
| chr4_+_72718458 | 5.45 |
ENSRNOT00000044780
|
Arhgef5
|
Rho guanine nucleotide exchange factor 5 |
| chr14_+_45062662 | 5.35 |
ENSRNOT00000059124
|
Tlr10
|
toll-like receptor 10 |
| chr7_-_119797098 | 5.27 |
ENSRNOT00000009994
|
Rac2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr2_-_168260536 | 5.10 |
ENSRNOT00000072551
|
Vom1r55
|
vomeronasal 1 receptor 55 |
| chr1_+_229107911 | 5.09 |
ENSRNOT00000016484
|
Olr338
|
olfactory receptor 338 |
| chr4_+_156752082 | 5.09 |
ENSRNOT00000084588
ENSRNOT00000068407 |
Cd163
|
CD163 molecule |
| chr2_-_80667481 | 4.97 |
ENSRNOT00000016784
|
Trio
|
trio Rho guanine nucleotide exchange factor |
| chr11_-_61470348 | 4.96 |
ENSRNOT00000083841
|
LOC102553099
|
N-alpha-acetyltransferase 50-like |
| chr19_+_15195565 | 4.78 |
ENSRNOT00000090865
ENSRNOT00000078874 |
Ces1d
|
carboxylesterase 1D |
| chr7_+_117643206 | 4.69 |
ENSRNOT00000077588
|
Adck5
|
aarF domain containing kinase 5 |
| chr4_+_66276835 | 4.51 |
ENSRNOT00000007544
|
Fmc1
|
formation of mitochondrial complex V assembly factor 1 |
| chr1_-_77893509 | 4.50 |
ENSRNOT00000015059
|
Gltscr1
|
glioma tumor suppressor candidate region gene 1 |
| chr13_+_113373578 | 4.45 |
ENSRNOT00000009900
|
Plxna2
|
plexin A2 |
| chr1_-_214252456 | 4.44 |
ENSRNOT00000023504
|
Irf7
|
interferon regulatory factor 7 |
| chr4_-_164453171 | 4.42 |
ENSRNOT00000077539
ENSRNOT00000083610 ENSRNOT00000079975 |
Ly49s6
|
Ly49 stimulatory receptor 6 |
| chr3_-_112614503 | 4.37 |
ENSRNOT00000071591
|
LOC100911313
|
regulator of microtubule dynamics protein 3-like |
| chr18_+_57516347 | 4.35 |
ENSRNOT00000082215
|
Gm9949
|
predicted gene 9949 |
| chr2_-_193136520 | 4.33 |
ENSRNOT00000042142
|
Kprp
|
keratinocyte proline-rich protein |
| chr10_-_14848967 | 4.27 |
ENSRNOT00000080944
|
Sstr5
|
somatostatin receptor 5 |
| chr11_+_73936750 | 4.15 |
ENSRNOT00000002350
|
Atp13a3
|
ATPase 13A3 |
| chr1_+_88686731 | 4.09 |
ENSRNOT00000028222
|
Polr2i
|
RNA polymerase II subunit I |
| chr1_+_79289164 | 4.00 |
ENSRNOT00000024155
|
AABR07002659.1
|
|
| chr17_-_67945037 | 3.92 |
ENSRNOT00000023033
|
Klf6
|
Kruppel-like factor 6 |
| chr5_+_173148884 | 3.83 |
ENSRNOT00000041753
|
LOC100364191
|
hCG1994130-like |
| chr2_-_210116038 | 3.63 |
ENSRNOT00000074950
|
LOC684509
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr2_+_211159062 | 3.62 |
ENSRNOT00000027099
|
Mybphl
|
myosin binding protein H-like |
| chr5_-_160423811 | 3.61 |
ENSRNOT00000018864
|
Efhd2
|
EF-hand domain family, member D2 |
| chr7_-_60860990 | 3.47 |
ENSRNOT00000009511
|
Rap1b
|
RAP1B, member of RAS oncogene family |
| chr2_-_187786700 | 3.47 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr12_-_16201632 | 3.46 |
ENSRNOT00000001688
|
Chst12
|
carbohydrate sulfotransferase 12 |
| chr7_-_9104719 | 3.43 |
ENSRNOT00000088247
|
LOC103692758
|
olfactory receptor 6C74-like |
| chr1_-_91042230 | 3.26 |
ENSRNOT00000073107
|
LOC103690054
|
DNA-directed RNA polymerase II subunit RPB9 |
| chr4_-_119568736 | 3.10 |
ENSRNOT00000041234
|
Aplf
|
aprataxin and PNKP like factor |
| chr3_-_59166356 | 3.01 |
ENSRNOT00000047713
|
AABR07052519.1
|
|
| chr2_+_78245459 | 2.97 |
ENSRNOT00000089551
|
LOC103689968
|
protein FAM134B |
| chr6_-_140216072 | 2.92 |
ENSRNOT00000072365
|
AABR07065750.1
|
|
| chr7_+_9279620 | 2.77 |
ENSRNOT00000011225
|
Olr1064
|
olfactory receptor 1064 |
| chr1_-_214159157 | 2.74 |
ENSRNOT00000091292
ENSRNOT00000022241 |
Rnh1
|
ribonuclease/angiogenin inhibitor 1 |
| chr9_-_11108741 | 2.70 |
ENSRNOT00000072357
|
Ccdc94
|
coiled-coil domain containing 94 |
| chr6_+_96834525 | 2.60 |
ENSRNOT00000077935
|
Hif1a
|
hypoxia inducible factor 1 alpha subunit |
| chr1_+_68239314 | 2.59 |
ENSRNOT00000070823
|
Vom1r109
|
vomeronasal 1 receptor 109 |
| chr14_+_18983853 | 2.57 |
ENSRNOT00000003836
|
Rassf6
|
Ras association domain family member 6 |
| chr1_-_169386939 | 2.54 |
ENSRNOT00000023007
|
Olr150
|
olfactory receptor 150 |
| chr4_+_45034123 | 2.46 |
ENSRNOT00000078255
|
ST7
|
suppression of tumorigenicity 7 |
| chr2_-_195935878 | 2.42 |
ENSRNOT00000028440
|
Cgn
|
cingulin |
| chr1_+_91042635 | 2.35 |
ENSRNOT00000028211
|
LOC103690005
|
tubulin-folding cofactor B |
| chr3_-_152179193 | 2.33 |
ENSRNOT00000026700
|
Rbm12
|
RNA binding motif protein 12 |
| chr1_+_172807389 | 2.31 |
ENSRNOT00000088537
|
Olr271
|
olfactory receptor 271 |
| chr19_+_38422164 | 2.30 |
ENSRNOT00000017174
|
Nqo1
|
NAD(P)H quinone dehydrogenase 1 |
| chr15_+_28295368 | 2.29 |
ENSRNOT00000013786
|
Slc39a2
|
solute carrier family 39 member 2 |
| chr1_-_225830300 | 2.28 |
ENSRNOT00000072584
ENSRNOT00000027502 |
Scgb2a2
|
secretoglobin, family 2A, member 2 |
| chr6_-_115352681 | 2.18 |
ENSRNOT00000005873
|
Gtf2a1
|
general transcription factor 2A subunit 1 |
| chr8_+_32604365 | 2.11 |
ENSRNOT00000071210
|
AABR07069587.1
|
|
| chr8_-_76940094 | 2.07 |
ENSRNOT00000082709
ENSRNOT00000084313 |
Rnf111
|
ring finger protein 111 |
| chr14_-_25585222 | 2.06 |
ENSRNOT00000042106
|
RGD1565048
|
similar to 60S ribosomal protein L9 |
| chr16_+_7758996 | 2.06 |
ENSRNOT00000061063
|
Btd
|
biotinidase |
| chr13_-_84331905 | 2.06 |
ENSRNOT00000004965
|
Dusp27
|
dual specificity phosphatase 27 (putative) |
| chr10_+_17261541 | 2.04 |
ENSRNOT00000005478
|
Sh3pxd2b
|
SH3 and PX domains 2B |
| chr3_+_73425801 | 2.04 |
ENSRNOT00000040714
|
Olr477
|
olfactory receptor 477 |
| chr6_+_137323713 | 2.02 |
ENSRNOT00000029017
|
Pld4
|
phospholipase D family, member 4 |
| chr4_-_68832216 | 1.99 |
ENSRNOT00000031345
|
Tas2r138
|
taste receptor, type 2, member 138 |
| chr3_-_77494108 | 1.91 |
ENSRNOT00000047726
|
Olr662
|
olfactory receptor 662 |
| chr9_-_85617954 | 1.85 |
ENSRNOT00000077331
|
Serpine2
|
serpin family E member 2 |
| chr3_+_78086943 | 1.84 |
ENSRNOT00000047425
|
Olr691
|
olfactory receptor 691 |
| chr7_-_10430992 | 1.83 |
ENSRNOT00000047433
|
Zfp709l1
|
zinc finger protein 709-like 1 |
| chr8_+_20230082 | 1.82 |
ENSRNOT00000044463
|
Olr1165
|
olfactory receptor 1165 |
| chrX_+_9956370 | 1.75 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr4_+_72428128 | 1.75 |
ENSRNOT00000007157
|
Olr811
|
olfactory receptor 811 |
| chr9_+_23379200 | 1.69 |
ENSRNOT00000074110
|
Glyatl3
|
glycine-N-acyltransferase-like 3 |
| chr1_+_68176904 | 1.68 |
ENSRNOT00000044950
|
Vom1r28
|
vomeronasal 1 receptor 28 |
| chr19_+_34616768 | 1.64 |
ENSRNOT00000051194
|
Rps27a-ps6
|
ribosomal protein S27a, pseudogene 6 |
| chr2_+_35935670 | 1.62 |
ENSRNOT00000076875
ENSRNOT00000075753 |
LOC108348103
|
serine protease inhibitor Kazal-type 5-like |
| chr18_+_35121967 | 1.56 |
ENSRNOT00000017522
|
Spink5
|
serine peptidase inhibitor, Kazal type 5 |
| chr5_-_64622360 | 1.54 |
ENSRNOT00000073545
|
Olr841
|
olfactory receptor 841 |
| chr11_+_43194348 | 1.53 |
ENSRNOT00000075303
|
Olr1532
|
olfactory receptor 1532 |
| chr3_-_145641654 | 1.49 |
ENSRNOT00000008101
ENSRNOT00000008047 |
LOC102554315
|
zinc finger protein 596-like |
| chr1_+_172813394 | 1.49 |
ENSRNOT00000090059
|
Olr272
|
olfactory receptor 272 |
| chr1_+_229267916 | 1.45 |
ENSRNOT00000073717
ENSRNOT00000082670 ENSRNOT00000076941 |
LOC100910851
|
serine protease inhibitor Kazal-type 5-like |
| chr8_+_133210473 | 1.45 |
ENSRNOT00000082774
|
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr4_+_170518673 | 1.41 |
ENSRNOT00000011803
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr10_+_31813814 | 1.40 |
ENSRNOT00000009573
|
Havcr1
|
hepatitis A virus cellular receptor 1 |
| chrX_-_102039530 | 1.40 |
ENSRNOT00000085348
|
AABR07040480.1
|
|
| chr1_+_57692836 | 1.39 |
ENSRNOT00000083968
ENSRNOT00000019358 |
Chd1
|
chromodomain helicase DNA binding protein 1 |
| chr10_+_60849405 | 1.39 |
ENSRNOT00000066983
|
LOC687896
|
hypothetical protein LOC687896 |
| chr10_+_14656812 | 1.38 |
ENSRNOT00000024904
|
Prss34
|
protease, serine, 34 |
| chr5_-_155728300 | 1.37 |
ENSRNOT00000029025
|
Cdc42
|
cell division cycle 42 |
| chr18_+_35384743 | 1.35 |
ENSRNOT00000076143
ENSRNOT00000074593 |
LOC100911797
|
serine protease inhibitor Kazal-type 5-like |
| chr7_+_114997103 | 1.33 |
ENSRNOT00000010224
|
Ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr4_-_121969446 | 1.33 |
ENSRNOT00000085001
|
Vom1r99
|
vomeronasal 1 receptor 99 |
| chr6_+_64224861 | 1.30 |
ENSRNOT00000093159
ENSRNOT00000093664 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr20_-_4818568 | 1.30 |
ENSRNOT00000001115
|
Ddx39b
|
DExD-box helicase 39B |
| chr1_-_71909138 | 1.27 |
ENSRNOT00000029591
|
Nlrp9
|
NLR family, pyrin domain containing 9 |
| chr13_+_9464906 | 1.26 |
ENSRNOT00000071756
|
LOC100910928
|
olfactory receptor 6C74-like |
| chr2_+_26240385 | 1.26 |
ENSRNOT00000024292
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr8_+_40258985 | 1.25 |
ENSRNOT00000044652
|
Olr1196
|
olfactory receptor 1196 |
| chr10_+_36477692 | 1.24 |
ENSRNOT00000044464
|
Olr1405
|
olfactory receptor 1405 |
| chr1_+_87045335 | 1.21 |
ENSRNOT00000084393
|
Lgals7
|
galectin 7 |
| chr1_-_228014924 | 1.16 |
ENSRNOT00000042091
|
Oosp2
|
oocyte secreted protein 2 |
| chr5_-_64615714 | 1.10 |
ENSRNOT00000007462
|
LOC500460
|
similar to olfactory receptor Olr841 |
| chr1_+_168489077 | 1.10 |
ENSRNOT00000021191
|
Olr96
|
olfactory receptor 96 |
| chr13_-_34253910 | 1.08 |
ENSRNOT00000092847
|
Tsn
|
translin |
| chr10_-_104689178 | 1.05 |
ENSRNOT00000011328
|
Mrpl38
|
mitochondrial ribosomal protein L38 |
| chr3_+_158964146 | 1.05 |
ENSRNOT00000073941
|
LOC100912340
|
olfactory receptor 8B8-like |
| chr3_-_102826379 | 1.01 |
ENSRNOT00000073423
|
Olr769
|
olfactory receptor 769 |
| chr1_-_60828260 | 1.00 |
ENSRNOT00000059671
|
Vom1r-ps112
|
vomeronasal 1 receptor pseudogene 112 |
| chr3_-_78595778 | 1.00 |
ENSRNOT00000084255
|
Olr715
|
olfactory receptor 715 |
| chr8_-_37093082 | 0.99 |
ENSRNOT00000072767
|
Pate-f
|
prostate and testis expressed protein F |
| chr1_+_87045796 | 0.91 |
ENSRNOT00000027620
|
Lgals7
|
galectin 7 |
| chrX_+_19854836 | 0.90 |
ENSRNOT00000075010
|
AABR07037395.2
|
|
| chr8_-_43055175 | 0.89 |
ENSRNOT00000074413
|
LOC103693054
|
olfactory receptor 8G1-like |
| chr3_+_21276213 | 0.87 |
ENSRNOT00000041835
|
Olr428
|
olfactory receptor 428 |
| chr8_+_69753373 | 0.86 |
ENSRNOT00000066973
|
Tipin
|
timeless interacting protein |
| chr1_-_168587241 | 0.85 |
ENSRNOT00000021254
|
Olr104
|
olfactory receptor 104 |
| chrX_+_77076106 | 0.83 |
ENSRNOT00000091527
ENSRNOT00000089381 |
Atp7a
|
ATPase copper transporting alpha |
| chr8_-_21453190 | 0.83 |
ENSRNOT00000078192
|
Zfp26
|
zinc finger protein 26 |
| chrX_+_23416395 | 0.82 |
ENSRNOT00000089821
|
AABR07037545.1
|
|
| chr13_-_111948753 | 0.75 |
ENSRNOT00000048074
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr11_+_73693814 | 0.75 |
ENSRNOT00000081081
ENSRNOT00000002354 ENSRNOT00000090940 |
Lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr8_+_42409187 | 0.74 |
ENSRNOT00000072605
|
Olr1256
|
olfactory receptor 1256 |
| chr1_-_167884690 | 0.74 |
ENSRNOT00000091372
|
Olr61
|
olfactory receptor 61 |
| chr18_-_68934953 | 0.73 |
ENSRNOT00000058894
|
AABR07032503.1
|
|
| chr15_+_35942770 | 0.73 |
ENSRNOT00000072814
|
Olr1278
|
olfactory receptor 1278 |
| chr12_-_16126953 | 0.72 |
ENSRNOT00000001682
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr5_+_59326803 | 0.71 |
ENSRNOT00000052426
|
Olr840
|
olfactory receptor 840 |
| chr3_-_113091770 | 0.67 |
ENSRNOT00000068132
|
Lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr16_-_81714346 | 0.67 |
ENSRNOT00000092552
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chrX_-_77418525 | 0.61 |
ENSRNOT00000092303
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr1_-_171162912 | 0.59 |
ENSRNOT00000074866
|
Olr229
|
olfactory receptor 229 |
| chr10_+_62566732 | 0.58 |
ENSRNOT00000021368
|
Taok1
|
TAO kinase 1 |
| chr17_+_77195247 | 0.54 |
ENSRNOT00000081940
|
Optn
|
optineurin |
| chr6_-_127571016 | 0.54 |
ENSRNOT00000057322
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chr7_+_10937599 | 0.50 |
ENSRNOT00000008758
|
Sirt6
|
sirtuin 6 |
| chr7_-_11195866 | 0.50 |
ENSRNOT00000005640
|
Dohh
|
deoxyhypusine hydroxylase/monooxygenase |
| chr8_-_37026142 | 0.46 |
ENSRNOT00000060446
|
LOC100361828
|
rCG22807-like |
| chrX_+_105575759 | 0.36 |
ENSRNOT00000088760
|
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr8_+_19289506 | 0.35 |
ENSRNOT00000044135
|
Olr1143
|
olfactory receptor 1143 |
| chr1_+_230160365 | 0.31 |
ENSRNOT00000088190
|
Olr361
|
olfactory receptor 361 |
| chr1_-_149529350 | 0.29 |
ENSRNOT00000052226
|
Vom2r43
|
vomeronasal 2 receptor, 43 |
| chr4_-_150520363 | 0.27 |
ENSRNOT00000065579
|
Zfp9
|
zinc finger protein 9 |
| chr1_-_169770188 | 0.27 |
ENSRNOT00000072602
|
Olr165
|
olfactory receptor 165 |
| chr14_+_70780623 | 0.23 |
ENSRNOT00000083871
ENSRNOT00000058803 |
Ldb2
|
LIM domain binding 2 |
| chr2_-_211690716 | 0.18 |
ENSRNOT00000027690
|
Fndc7
|
fibronectin type III domain containing 7 |
| chr5_+_146294030 | 0.17 |
ENSRNOT00000073637
|
AABR07049960.1
|
|
| chr1_+_72280117 | 0.14 |
ENSRNOT00000021077
|
Rfpl4a
|
ret finger protein-like 4A |
| chr12_+_17253791 | 0.00 |
ENSRNOT00000083814
|
Zfand2a
|
zinc finger AN1-type containing 2A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Six3_Six1_Six2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.4 | 37.3 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 10.0 | 39.9 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 9.9 | 39.4 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 5.0 | 15.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 4.8 | 14.4 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 4.4 | 13.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 3.2 | 22.5 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 3.0 | 26.9 | GO:0019530 | allantoin metabolic process(GO:0000255) isoleucine metabolic process(GO:0006549) creatine metabolic process(GO:0006600) taurine metabolic process(GO:0019530) creatinine metabolic process(GO:0046449) |
| 2.7 | 21.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 2.2 | 6.7 | GO:0032741 | central nervous system myelin formation(GO:0032289) positive regulation of interleukin-18 production(GO:0032741) detection of diacyl bacterial lipopeptide(GO:0042496) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) detection of bacterial lipopeptide(GO:0070340) |
| 2.2 | 10.9 | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste(GO:0001582) |
| 1.7 | 28.7 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 1.7 | 26.9 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 1.6 | 4.8 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 1.3 | 2.6 | GO:0080033 | cellular response to nitrite(GO:0071250) response to nitrite(GO:0080033) |
| 1.2 | 3.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 1.1 | 5.4 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.9 | 7.3 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.8 | 3.1 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.7 | 4.4 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.7 | 4.3 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.7 | 3.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.7 | 1.4 | GO:0071338 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.7 | 5.3 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.7 | 3.9 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.6 | 3.0 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.5 | 6.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 4.4 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.5 | 1.4 | GO:2000464 | positive regulation of astrocyte chemotaxis(GO:2000464) |
| 0.5 | 12.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 1.8 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.3 | 2.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) positive regulation of adipose tissue development(GO:1904179) |
| 0.3 | 1.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.3 | 4.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 2.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 | 1.4 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.3 | 1.6 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.3 | 0.8 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.2 | 7.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 7.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.2 | 0.7 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.2 | 0.8 | GO:1904959 | negative regulation of iron ion transmembrane transport(GO:0034760) elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.2 | 0.7 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.2 | 7.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.2 | 1.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 16.8 | GO:0044264 | cellular polysaccharide metabolic process(GO:0044264) |
| 0.2 | 8.2 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 1.3 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 0.9 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.5 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.1 | 1.3 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 6.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 6.0 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 1.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 1.4 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 1.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 2.3 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 2.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 2.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 5.9 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 1.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.5 | GO:0061734 | negative regulation of receptor recycling(GO:0001920) parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.1 | 2.1 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 2.4 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 61.4 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 2.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 6.8 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 5.1 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 1.9 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.2 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 2.7 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 26.9 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 2.2 | 6.7 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 2.2 | 10.9 | GO:1903768 | sweet taste receptor complex(GO:1903767) taste receptor complex(GO:1903768) |
| 1.9 | 17.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.8 | 9.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.4 | 6.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.4 | 2.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 1.6 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 26.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 40.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 4.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 1.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 0.7 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.2 | 1.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 3.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 6.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 19.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 8.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 7.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.5 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.9 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 3.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 92.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 3.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 3.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 5.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 2.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 2.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 4.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 7.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 75.1 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 3.2 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 18.6 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 37.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 7.5 | 22.5 | GO:0050656 | steroid sulfotransferase activity(GO:0050294) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 6.7 | 26.9 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 6.6 | 39.4 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 4.2 | 21.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 2.7 | 10.9 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 2.2 | 6.7 | GO:0042498 | diacyl lipopeptide binding(GO:0042498) |
| 2.0 | 12.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.8 | 28.7 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 1.2 | 4.8 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.9 | 3.5 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.7 | 9.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.7 | 4.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.7 | 2.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.7 | 7.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.5 | 15.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.5 | 14.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.5 | 1.4 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 0.5 | 2.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.5 | 1.4 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.5 | 2.7 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.4 | 6.9 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.4 | 8.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.4 | 39.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.3 | 4.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 1.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 1.7 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.3 | 2.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 2.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.3 | 0.8 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 12.9 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.2 | 6.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 3.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 7.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.2 | 0.8 | GO:0032767 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 1.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 1.3 | GO:0017070 | U6 snRNA binding(GO:0017070) U4 snRNA binding(GO:0030621) |
| 0.1 | 2.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 11.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.5 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 0.1 | 16.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 11.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 30.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 5.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 8.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 2.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 4.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 22.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 3.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 2.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 27.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.7 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 5.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.2 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 1.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 10.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 3.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 29.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.4 | 20.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 21.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 6.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 5.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 6.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 3.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 1.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 16.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 5.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 39.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 2.0 | 28.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.7 | 22.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 1.7 | 6.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 1.3 | 26.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 1.1 | 13.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.1 | 21.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 1.0 | 37.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.6 | 10.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.5 | 15.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.5 | 4.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.4 | 6.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.4 | 4.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 1.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.3 | 4.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.3 | 6.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 3.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 5.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 2.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 6.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 5.3 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 2.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 2.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 4.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |