Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
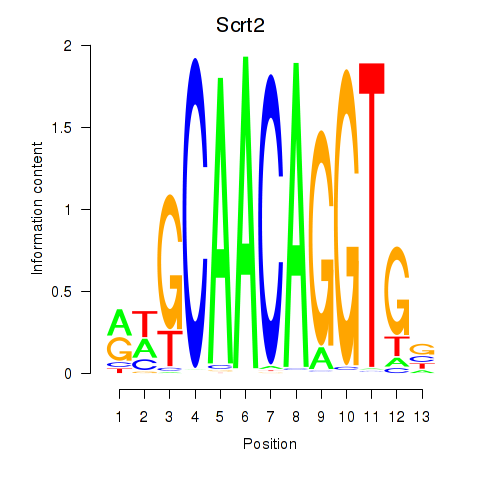
Results for Scrt2
Z-value: 1.05
Transcription factors associated with Scrt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Scrt2
|
ENSRNOG00000005148 | scratch family transcriptional repressor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Scrt2 | rn6_v1_chr3_+_147585947_147585947 | 0.57 | 4.5e-29 | Click! |
Activity profile of Scrt2 motif
Sorted Z-values of Scrt2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_49676540 | 59.92 |
ENSRNOT00000022032
ENSRNOT00000082205 |
Fxyd6
|
FXYD domain-containing ion transport regulator 6 |
| chr2_-_119537837 | 53.94 |
ENSRNOT00000015200
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr14_-_43143973 | 53.21 |
ENSRNOT00000003248
|
Uchl1
|
ubiquitin C-terminal hydrolase L1 |
| chr2_+_121165137 | 47.48 |
ENSRNOT00000016236
|
Sox2
|
SRY box 2 |
| chr9_-_85243001 | 46.52 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr18_-_6782996 | 42.93 |
ENSRNOT00000090320
|
Aqp4
|
aquaporin 4 |
| chr18_-_6782757 | 42.77 |
ENSRNOT00000068150
|
Aqp4
|
aquaporin 4 |
| chr1_-_93949187 | 41.76 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr10_+_58332588 | 38.46 |
ENSRNOT00000081954
|
Wscd1
|
WSC domain containing 1 |
| chrX_+_17171605 | 37.77 |
ENSRNOT00000048236
|
Nudt10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
| chr7_+_121408829 | 35.36 |
ENSRNOT00000023434
|
Mgat3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr14_-_78308933 | 35.31 |
ENSRNOT00000065334
|
Crmp1
|
collapsin response mediator protein 1 |
| chr4_-_150829913 | 35.09 |
ENSRNOT00000041571
|
Cacna1c
|
calcium voltage-gated channel subunit alpha1 C |
| chr3_-_143063983 | 35.06 |
ENSRNOT00000006329
|
Napb
|
NSF attachment protein beta |
| chr10_+_91710495 | 33.21 |
ENSRNOT00000033276
|
Rprml
|
reprimo-like |
| chr4_-_150829741 | 33.18 |
ENSRNOT00000051846
ENSRNOT00000052017 |
Cacna1c
|
calcium voltage-gated channel subunit alpha1 C |
| chr8_+_64481172 | 33.14 |
ENSRNOT00000015332
|
Pkm
|
pyruvate kinase, muscle |
| chr20_+_37876650 | 30.80 |
ENSRNOT00000001054
|
Gja1
|
gap junction protein, alpha 1 |
| chr1_-_199624783 | 26.87 |
ENSRNOT00000026908
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr1_+_106896790 | 26.23 |
ENSRNOT00000042952
ENSRNOT00000064193 |
Ano5
|
anoctamin 5 |
| chr2_-_9504134 | 26.18 |
ENSRNOT00000076996
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr4_-_83972540 | 25.34 |
ENSRNOT00000036951
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr2_+_28049217 | 24.23 |
ENSRNOT00000022144
|
Enc1
|
ectodermal-neural cortex 1 |
| chr20_-_47306318 | 22.69 |
ENSRNOT00000075151
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr8_+_4440876 | 22.47 |
ENSRNOT00000049325
ENSRNOT00000076529 ENSRNOT00000076748 |
Pdgfd
|
platelet derived growth factor D |
| chr15_+_67555835 | 22.43 |
ENSRNOT00000045882
|
Pcdh17
|
protocadherin 17 |
| chr1_+_266953139 | 21.20 |
ENSRNOT00000054696
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr12_-_44381289 | 20.59 |
ENSRNOT00000001493
|
Nos1
|
nitric oxide synthase 1 |
| chr15_+_33544312 | 20.44 |
ENSRNOT00000020409
|
Bcl2l2
|
Bcl2-like 2 |
| chr11_-_88972176 | 20.14 |
ENSRNOT00000002498
|
Pkp2
|
plakophilin 2 |
| chrX_-_140303686 | 18.54 |
ENSRNOT00000033219
|
Gpr101
|
G protein-coupled receptor 101 |
| chr12_+_2534212 | 18.19 |
ENSRNOT00000001399
|
Ctxn1
|
cortexin 1 |
| chr13_-_42263024 | 17.60 |
ENSRNOT00000004741
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr20_+_5050327 | 16.73 |
ENSRNOT00000083353
|
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr18_+_39172028 | 16.37 |
ENSRNOT00000086651
|
Kcnn2
|
potassium calcium-activated channel subfamily N member 2 |
| chrX_-_23649466 | 15.84 |
ENSRNOT00000059431
|
Shroom2
|
shroom family member 2 |
| chr2_-_142885604 | 14.44 |
ENSRNOT00000031487
|
Frem2
|
Fras1 related extracellular matrix protein 2 |
| chr7_-_59514939 | 12.88 |
ENSRNOT00000085579
|
Kcnmb4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr10_-_40764185 | 12.76 |
ENSRNOT00000017486
|
Sparc
|
secreted protein acidic and cysteine rich |
| chr4_+_34962339 | 12.74 |
ENSRNOT00000086248
|
AABR07059778.1
|
|
| chr5_+_28480023 | 12.41 |
ENSRNOT00000075067
ENSRNOT00000093754 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr7_+_94130852 | 11.87 |
ENSRNOT00000011485
|
Mal2
|
mal, T-cell differentiation protein 2 |
| chr10_+_11912543 | 11.11 |
ENSRNOT00000045192
|
Zfp597
|
zinc finger protein 597 |
| chr16_+_8128689 | 11.08 |
ENSRNOT00000065618
|
Galnt15
|
polypeptide N-acetylgalactosaminyltransferase 15 |
| chrX_+_134742356 | 11.03 |
ENSRNOT00000005267
ENSRNOT00000082363 |
Ocrl
|
OCRL, inositol polyphosphate-5-phosphatase |
| chr1_-_54748763 | 10.24 |
ENSRNOT00000074549
|
LOC100911027
|
protein MAL2-like |
| chr2_-_227160379 | 10.10 |
ENSRNOT00000066581
|
Usp53
|
ubiquitin specific peptidase 53 |
| chrX_-_70978714 | 9.73 |
ENSRNOT00000076336
|
Slc7a3
|
solute carrier family 7 member 3 |
| chr1_-_167911961 | 9.36 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr3_+_100768637 | 9.19 |
ENSRNOT00000083542
|
Bdnf
|
brain-derived neurotrophic factor |
| chr1_-_15301998 | 9.01 |
ENSRNOT00000016400
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr7_+_12974169 | 8.84 |
ENSRNOT00000010555
|
C2cd4c
|
C2 calcium-dependent domain containing 4C |
| chr4_+_158243086 | 7.48 |
ENSRNOT00000032112
|
Ano2
|
anoctamin 2 |
| chr13_+_50873605 | 7.47 |
ENSRNOT00000004382
|
Fmod
|
fibromodulin |
| chr2_-_210874304 | 7.18 |
ENSRNOT00000088657
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr18_+_65285318 | 6.78 |
ENSRNOT00000020431
|
Tcf4
|
transcription factor 4 |
| chr12_+_38345456 | 6.70 |
ENSRNOT00000001685
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr10_-_15211325 | 6.53 |
ENSRNOT00000027083
|
Rhot2
|
ras homolog family member T2 |
| chr2_-_188413219 | 6.19 |
ENSRNOT00000065065
|
Fdps
|
farnesyl diphosphate synthase |
| chr15_-_28406046 | 5.75 |
ENSRNOT00000015418
|
Zfp219
|
zinc finger protein 219 |
| chrX_+_53360839 | 5.54 |
ENSRNOT00000091467
ENSRNOT00000034372 ENSRNOT00000081061 |
Dmd
|
dystrophin |
| chr18_-_79258570 | 5.51 |
ENSRNOT00000022401
|
Galr1
|
galanin receptor 1 |
| chr13_+_101698096 | 5.21 |
ENSRNOT00000089309
|
Aida
|
axin interactor, dorsalization associated |
| chr8_-_28208466 | 5.18 |
ENSRNOT00000012247
|
Jam3
|
junctional adhesion molecule 3 |
| chr1_+_66004349 | 4.85 |
ENSRNOT00000042276
ENSRNOT00000052029 |
Vom2r34
|
vomeronasal 2 receptor, 34 |
| chr7_+_5593735 | 4.45 |
ENSRNOT00000042179
|
Olr917
|
olfactory receptor 917 |
| chr7_+_59349745 | 4.44 |
ENSRNOT00000085334
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr1_+_171592797 | 3.82 |
ENSRNOT00000026607
|
Syt9
|
synaptotagmin 9 |
| chr1_+_193335645 | 3.76 |
ENSRNOT00000019640
|
Lcmt1
|
leucine carboxyl methyltransferase 1 |
| chrX_-_124576133 | 3.62 |
ENSRNOT00000041904
|
Rhox3
|
reproductive homeobox on X chromosome 3 |
| chr10_+_74886985 | 3.38 |
ENSRNOT00000009943
|
Mtmr4
|
myotubularin related protein 4 |
| chr1_-_188101566 | 3.38 |
ENSRNOT00000092435
|
Syt17
|
synaptotagmin 17 |
| chr1_+_53220397 | 3.14 |
ENSRNOT00000089989
|
AABR07001573.1
|
|
| chr10_+_55555089 | 2.91 |
ENSRNOT00000006597
|
Slc25a35
|
solute carrier family 25, member 35 |
| chr1_+_78820262 | 2.76 |
ENSRNOT00000022441
|
Gng8
|
G protein subunit gamma 8 |
| chrX_-_70978952 | 2.76 |
ENSRNOT00000076910
|
Slc7a3
|
solute carrier family 7 member 3 |
| chr1_-_79690434 | 2.75 |
ENSRNOT00000057986
ENSRNOT00000057988 |
LOC102557319
|
carcinoembryonic antigen-related cell adhesion molecule 3-like |
| chr1_-_253185533 | 2.57 |
ENSRNOT00000067822
|
Pank1
|
pantothenate kinase 1 |
| chr3_-_9262628 | 2.57 |
ENSRNOT00000013286
|
Aif1l
|
allograft inflammatory factor 1-like |
| chr5_-_78183122 | 1.73 |
ENSRNOT00000002083
ENSRNOT00000067076 |
Fkbp15
|
FK506 binding protein 15 |
| chr19_-_25261911 | 1.51 |
ENSRNOT00000045180
ENSRNOT00000009806 |
Cc2d1a
|
coiled-coil and C2 domain containing 1A |
| chr10_-_43895376 | 1.36 |
ENSRNOT00000031811
|
Olr1413
|
olfactory receptor 1413 |
| chr2_-_211638788 | 1.28 |
ENSRNOT00000027666
|
Stxbp3
|
syntaxin binding protein 3 |
| chr20_+_31102476 | 1.25 |
ENSRNOT00000078719
|
Lrrc20
|
leucine rich repeat containing 20 |
| chr7_-_90318221 | 1.22 |
ENSRNOT00000050774
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr3_-_11277757 | 1.21 |
ENSRNOT00000015792
|
Uck1
|
uridine-cytidine kinase 1 |
| chr11_+_46108470 | 1.02 |
ENSRNOT00000073467
|
Adgrg7
|
adhesion G protein-coupled receptor G7 |
| chr9_+_19451630 | 0.80 |
ENSRNOT00000065048
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr8_-_22874637 | 0.70 |
ENSRNOT00000064551
ENSRNOT00000090424 |
Dock6
|
dedicator of cytokinesis 6 |
| chr19_+_25123724 | 0.50 |
ENSRNOT00000007407
|
LOC686013
|
hypothetical protein LOC686013 |
| chr7_-_24667301 | 0.33 |
ENSRNOT00000009154
|
Tmem263
|
transmembrane protein 263 |
| chrX_-_123867149 | 0.30 |
ENSRNOT00000089754
|
Rhox3
|
reproductive homeobox on X chromosome 3 |
| chr7_+_122157201 | 0.23 |
ENSRNOT00000025193
|
Adsl
|
adenylosuccinate lyase |
| chr5_+_13379772 | 0.14 |
ENSRNOT00000010032
|
Npbwr1
|
neuropeptides B/W receptor 1 |
| chr12_+_22153983 | 0.00 |
ENSRNOT00000080775
ENSRNOT00000001894 |
Pcolce
|
procollagen C-endopeptidase enhancer |
Network of associatons between targets according to the STRING database.
First level regulatory network of Scrt2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.4 | 85.7 | GO:0060354 | negative regulation of cell adhesion molecule production(GO:0060354) |
| 11.9 | 47.5 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 11.4 | 68.3 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 10.3 | 30.8 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 9.4 | 37.8 | GO:0071543 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 8.8 | 35.3 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 8.8 | 35.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 7.7 | 53.9 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) positive regulation of corticotropin secretion(GO:0051461) |
| 7.5 | 22.5 | GO:2000437 | regulation of monocyte extravasation(GO:2000437) |
| 6.6 | 46.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 6.2 | 62.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 6.1 | 24.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 5.1 | 20.6 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 4.6 | 41.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 3.8 | 22.7 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 3.3 | 33.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 3.1 | 9.4 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 3.1 | 46.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 2.8 | 5.5 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 2.7 | 16.4 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 2.5 | 12.5 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 2.5 | 22.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 2.2 | 22.4 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 2.2 | 26.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.9 | 35.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 1.9 | 16.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 1.8 | 5.5 | GO:0021627 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 1.7 | 20.4 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 1.6 | 21.2 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 1.3 | 6.7 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 1.2 | 6.0 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) |
| 1.2 | 7.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.1 | 15.8 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 1.0 | 59.9 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.9 | 16.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.8 | 25.3 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.8 | 9.0 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.8 | 12.8 | GO:0009629 | response to gravity(GO:0009629) |
| 0.7 | 18.5 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.6 | 17.6 | GO:1903831 | acetylcholine receptor signaling pathway(GO:0095500) negative regulation of protein localization to plasma membrane(GO:1903077) signal transduction involved in cellular response to ammonium ion(GO:1903831) negative regulation of protein localization to cell periphery(GO:1904376) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.6 | 12.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.6 | 5.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.5 | 6.8 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.5 | 5.2 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.4 | 11.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.4 | 10.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.3 | 3.4 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 | 6.5 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.3 | 2.8 | GO:0043584 | nose development(GO:0043584) |
| 0.3 | 1.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.3 | 3.8 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.3 | 1.3 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.2 | 7.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 2.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 7.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 13.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 1.2 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 4.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.8 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 14.4 | GO:0007507 | heart development(GO:0007507) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 35.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 5.7 | 68.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 5.1 | 25.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 4.9 | 53.9 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 4.4 | 26.2 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 3.2 | 12.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 3.1 | 50.3 | GO:0031045 | dense core granule(GO:0031045) |
| 2.9 | 20.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 2.4 | 85.7 | GO:0046930 | pore complex(GO:0046930) |
| 1.7 | 26.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.6 | 30.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 1.1 | 20.6 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 1.1 | 51.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 1.0 | 53.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.9 | 20.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.8 | 6.7 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.8 | 21.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.6 | 5.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.6 | 9.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.4 | 1.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.4 | 16.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.4 | 11.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 24.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 5.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 47.5 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.3 | 6.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 33.1 | GO:0061695 | transferase complex, transferring phosphorus-containing groups(GO:0061695) |
| 0.1 | 14.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 112.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 22.1 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 2.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 4.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 6.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 7.5 | GO:0044431 | Golgi apparatus part(GO:0044431) |
| 0.0 | 1.5 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.3 | 53.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 11.7 | 35.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 11.4 | 68.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 10.8 | 53.9 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 9.4 | 37.8 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 8.3 | 33.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 6.2 | 30.8 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 6.1 | 85.7 | GO:0015250 | water channel activity(GO:0015250) |
| 5.6 | 16.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 5.2 | 41.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 4.1 | 20.6 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 3.0 | 47.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 2.3 | 16.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 2.3 | 6.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 2.1 | 12.5 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 1.8 | 22.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.8 | 7.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.8 | 35.3 | GO:0031005 | filamin binding(GO:0031005) |
| 1.7 | 33.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 1.6 | 44.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 1.5 | 20.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 1.4 | 11.0 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 1.3 | 59.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.3 | 20.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 1.3 | 17.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 1.2 | 18.5 | GO:0004935 | adrenergic receptor activity(GO:0004935) |
| 1.2 | 6.0 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 1.1 | 9.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.1 | 22.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 1.0 | 12.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 1.0 | 25.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.9 | 26.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.7 | 35.4 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.7 | 5.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.6 | 2.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.6 | 12.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.6 | 38.5 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.5 | 3.8 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.5 | 21.2 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.4 | 31.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.4 | 32.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.3 | 6.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 1.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 12.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 15.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 11.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.8 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 7.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 26.2 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 5.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 7.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 9.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 53.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.7 | 30.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.7 | 35.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.5 | 22.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 5.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 6.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 9.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 7.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 20.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 6.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 14.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 12.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 85.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 3.4 | 30.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 2.0 | 35.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 1.6 | 35.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 1.1 | 33.5 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.8 | 3.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.6 | 11.0 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.5 | 7.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 12.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 22.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 6.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 2.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 6.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 22.5 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 5.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.8 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.1 | 8.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 5.2 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 6.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 6.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 5.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |