Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
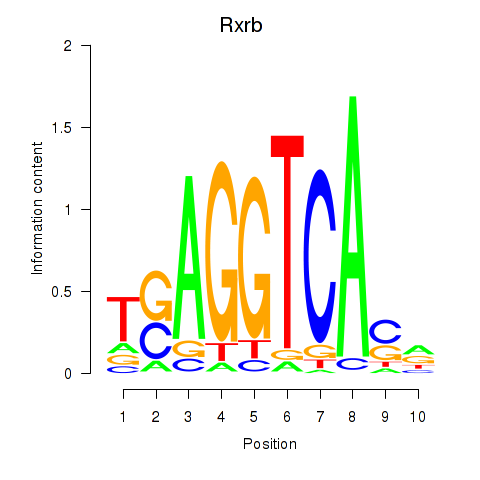
Results for Rxrb
Z-value: 0.85
Transcription factors associated with Rxrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rxrb
|
ENSRNOG00000000464 | retinoid X receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxrb | rn6_v1_chr20_+_3827367_3827367 | -0.13 | 2.2e-02 | Click! |
Activity profile of Rxrb motif
Sorted Z-values of Rxrb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_150052483 | 59.68 |
ENSRNOT00000022265
|
RGD1561517
|
similar to chromosome 20 open reading frame 144 |
| chr1_-_169334093 | 54.93 |
ENSRNOT00000032587
|
Ubqln3
|
ubiquilin 3 |
| chr10_-_82209459 | 54.73 |
ENSRNOT00000004377
|
Spata20
|
spermatogenesis associated 20 |
| chr13_-_74740458 | 51.76 |
ENSRNOT00000006548
|
Tex35
|
testis expressed 35 |
| chr7_+_77066955 | 49.76 |
ENSRNOT00000008700
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr19_-_42180981 | 48.46 |
ENSRNOT00000019577
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr7_+_117326279 | 45.33 |
ENSRNOT00000050522
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr1_-_169321075 | 44.68 |
ENSRNOT00000055216
|
RGD1562433
|
similar to ubiquilin 1 isoform 2 |
| chr3_-_149356160 | 43.10 |
ENSRNOT00000037589
ENSRNOT00000081775 |
Sun5
|
Sad1 and UNC84 domain containing 5 |
| chr1_+_72644333 | 41.65 |
ENSRNOT00000048976
ENSRNOT00000065051 |
Fam71e2
|
family with sequence similarity 71, member E2 |
| chr9_+_92435896 | 38.56 |
ENSRNOT00000022901
|
Fbxo36
|
F-box protein 36 |
| chr6_+_101288951 | 37.66 |
ENSRNOT00000046901
|
RGD1562540
|
RGD1562540 |
| chr9_+_10004805 | 37.04 |
ENSRNOT00000074117
|
Slc25a41
|
solute carrier family 25, member 41 |
| chr5_-_57372239 | 36.37 |
ENSRNOT00000012975
|
Aqp7
|
aquaporin 7 |
| chr15_+_2740784 | 36.28 |
ENSRNOT00000067781
|
Dusp13
|
dual specificity phosphatase 13 |
| chrX_+_13992064 | 36.24 |
ENSRNOT00000036543
|
LOC100363125
|
rCG42854-like |
| chr14_-_108284619 | 35.09 |
ENSRNOT00000078814
|
LOC690352
|
hypothetical protein LOC690352 |
| chr14_-_108285008 | 33.22 |
ENSRNOT00000087501
|
LOC690352
|
hypothetical protein LOC690352 |
| chr19_-_20508711 | 32.63 |
ENSRNOT00000032408
|
LOC680913
|
hypothetical protein LOC680913 |
| chr3_-_2434257 | 32.20 |
ENSRNOT00000013278
|
Stpg3
|
sperm-tail PG-rich repeat containing 3 |
| chr12_+_22142520 | 30.48 |
ENSRNOT00000038397
|
Fbxo24
|
F-box protein 24 |
| chr1_+_97712177 | 30.02 |
ENSRNOT00000027745
|
MGC114499
|
similar to RIKEN cDNA 4930433I11 gene |
| chr7_-_117326551 | 29.42 |
ENSRNOT00000086087
|
AC107096.1
|
|
| chr13_+_93751759 | 29.38 |
ENSRNOT00000065706
|
Wdr64
|
WD repeat domain 64 |
| chr1_+_72635267 | 29.34 |
ENSRNOT00000090636
|
Il11
|
interleukin 11 |
| chr11_-_32469566 | 28.60 |
ENSRNOT00000002718
|
LOC498063
|
similar to RIKEN cDNA 4930563D23 |
| chr5_-_75116490 | 28.50 |
ENSRNOT00000042788
|
Txndc8
|
thioredoxin domain containing 8 |
| chr20_+_4959294 | 28.32 |
ENSRNOT00000074223
|
Hspa1l
|
heat shock protein family A (Hsp70) member 1 like |
| chr4_-_6062641 | 27.68 |
ENSRNOT00000074846
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr5_-_136049551 | 27.66 |
ENSRNOT00000068395
ENSRNOT00000079371 |
Kif2c
|
kinesin family member 2C |
| chr10_-_55560422 | 26.58 |
ENSRNOT00000006883
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr3_-_134406576 | 25.15 |
ENSRNOT00000081589
|
Sel1l2
|
SEL1L2 ERAD E3 ligase adaptor subunit |
| chr6_-_136185651 | 23.28 |
ENSRNOT00000085594
|
Bag5
|
BCL2-associated athanogene 5 |
| chr6_-_136279399 | 22.47 |
ENSRNOT00000015333
|
Bag5l1
|
BCL2-associated athanogene 5-like 1 |
| chr1_+_97777685 | 15.12 |
ENSRNOT00000030895
|
LOC690284
|
similar to F49E2.5d |
| chr2_-_123851854 | 14.22 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr17_+_57040023 | 13.39 |
ENSRNOT00000020204
|
Crem
|
cAMP responsive element modulator |
| chr11_-_4332255 | 13.30 |
ENSRNOT00000087133
|
Cadm2
|
cell adhesion molecule 2 |
| chr5_+_168078748 | 12.78 |
ENSRNOT00000024798
|
Uts2
|
urotensin 2 |
| chr3_+_150055749 | 11.42 |
ENSRNOT00000055335
|
Actl10
|
actin-like 10 |
| chr14_-_37871051 | 8.81 |
ENSRNOT00000003087
|
Slain2
|
SLAIN motif family, member 2 |
| chr16_+_19044607 | 8.65 |
ENSRNOT00000085462
|
Med26
|
mediator complex subunit 26 |
| chr6_+_21051327 | 8.54 |
ENSRNOT00000052131
|
Fam98a
|
family with sequence similarity 98, member A |
| chr4_-_120390164 | 7.80 |
ENSRNOT00000017323
ENSRNOT00000063950 |
Eefsec
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr2_-_122646445 | 7.73 |
ENSRNOT00000080100
|
Dcun1d1
|
defective in cullin neddylation 1 domain containing 1 |
| chr5_+_169357517 | 7.12 |
ENSRNOT00000000079
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr6_-_122721496 | 6.41 |
ENSRNOT00000079697
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr4_-_157486844 | 5.77 |
ENSRNOT00000038281
ENSRNOT00000022874 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr9_-_115850634 | 5.15 |
ENSRNOT00000049720
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr2_+_198312428 | 4.22 |
ENSRNOT00000028758
|
Sf3b4
|
splicing factor 3b, subunit 4 |
| chr7_-_123586919 | 3.73 |
ENSRNOT00000011484
|
Ndufa6
|
NADH:ubiquinone oxidoreductase subunit A6 |
| chr4_+_57715946 | 2.95 |
ENSRNOT00000079059
ENSRNOT00000034429 |
Klhdc10
|
kelch domain containing 10 |
| chr12_+_38368693 | 2.93 |
ENSRNOT00000083261
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr1_-_266919031 | 2.69 |
ENSRNOT00000044519
|
Calhm1
|
calcium homeostasis modulator 1 |
| chr3_+_95715193 | 2.51 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr7_-_70480724 | 2.49 |
ENSRNOT00000090057
|
Dtx3
|
deltex E3 ubiquitin ligase 3 |
| chr9_-_44786041 | 2.32 |
ENSRNOT00000067197
|
Rev1
|
REV1, DNA directed polymerase |
| chr9_+_18564927 | 2.20 |
ENSRNOT00000061014
|
Runx2
|
runt-related transcription factor 2 |
| chr10_-_87521514 | 2.01 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr8_-_55087832 | 1.35 |
ENSRNOT00000032152
|
Dlat
|
dihydrolipoamide S-acetyltransferase |
| chr10_-_87535438 | 0.94 |
ENSRNOT00000086873
|
Krtap2-4
|
keratin associated protein 2-4 |
| chr2_+_118910587 | 0.77 |
ENSRNOT00000065518
|
Zfp639
|
zinc finger protein 639 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rxrb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.4 | 45.8 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 7.3 | 36.4 | GO:0015793 | glycerol transport(GO:0015793) |
| 5.4 | 43.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 4.6 | 27.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 4.1 | 37.0 | GO:0015866 | ADP transport(GO:0015866) |
| 3.7 | 29.3 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 3.6 | 14.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 3.0 | 26.6 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 2.6 | 12.8 | GO:0010841 | negative regulation of glomerular filtration(GO:0003105) positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 2.4 | 7.1 | GO:0036115 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) fatty-acyl-CoA catabolic process(GO:0036115) |
| 1.7 | 28.5 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 1.4 | 28.3 | GO:0042026 | protein refolding(GO:0042026) |
| 1.3 | 2.7 | GO:0015867 | ATP transport(GO:0015867) |
| 1.1 | 7.8 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.9 | 27.7 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.8 | 2.5 | GO:0003322 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.6 | 2.9 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.6 | 5.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.5 | 2.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.4 | 2.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.4 | 8.8 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.4 | 42.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.3 | 7.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.3 | 13.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 4.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 104.5 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 1.3 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 4.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.8 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 17.3 | GO:0007010 | cytoskeleton organization(GO:0007010) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 98.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 4.6 | 27.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 2.2 | 28.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 1.2 | 39.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 1.1 | 8.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.7 | 43.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.5 | 45.8 | GO:0016234 | inclusion body(GO:0016234) |
| 0.4 | 36.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 25.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.3 | 28.5 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.2 | 8.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 5.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 4.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 1.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 45.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 49.6 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.1 | 3.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 24.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 4.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.9 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.1 | 36.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 6.6 | 26.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 4.1 | 37.0 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 3.6 | 28.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 2.8 | 14.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 1.8 | 7.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 1.4 | 30.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 1.3 | 7.8 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 1.3 | 45.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.9 | 27.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.7 | 36.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.6 | 28.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.5 | 45.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.5 | 7.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 2.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 29.3 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.2 | 1.3 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 6.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 8.7 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 10.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 2.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 2.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 2.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 26.4 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.8 | GO:0043621 | protein self-association(GO:0043621) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 27.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.5 | 14.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 29.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 5.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 36.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.9 | 27.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 13.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 14.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 2.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 4.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 3.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 10.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.3 | REACTOME DNA REPAIR | Genes involved in DNA Repair |