Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
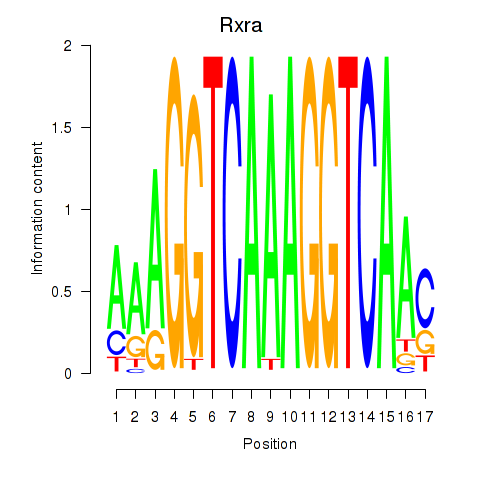
Results for Rxra
Z-value: 0.99
Transcription factors associated with Rxra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rxra
|
ENSRNOG00000009446 | retinoid X receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxra | rn6_v1_chr3_+_6211789_6211789 | -0.39 | 9.4e-13 | Click! |
Activity profile of Rxra motif
Sorted Z-values of Rxra motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_113318563 | 42.02 |
ENSRNOT00000089230
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr9_-_92530938 | 28.10 |
ENSRNOT00000064875
|
Slc16a14
|
solute carrier family 16, member 14 |
| chr2_-_225107283 | 27.95 |
ENSRNOT00000055711
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr17_+_23661429 | 26.49 |
ENSRNOT00000046523
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr4_-_176294997 | 24.58 |
ENSRNOT00000015112
ENSRNOT00000051461 ENSRNOT00000000026 ENSRNOT00000039877 ENSRNOT00000049303 |
Slc21a4
|
kidney specific organic anion transporter |
| chr10_-_65692016 | 24.33 |
ENSRNOT00000085074
ENSRNOT00000038690 |
Slc13a2
|
solute carrier family 13 member 2 |
| chr18_-_1946840 | 23.20 |
ENSRNOT00000041878
|
Abhd3
|
abhydrolase domain containing 3 |
| chr1_-_31122093 | 23.13 |
ENSRNOT00000016712
|
NEWGENE_1307525
|
SOGA family member 3 |
| chrX_+_14019961 | 22.93 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr7_+_139685573 | 22.92 |
ENSRNOT00000088376
|
Pfkm
|
phosphofructokinase, muscle |
| chr9_-_71852113 | 22.90 |
ENSRNOT00000083263
ENSRNOT00000072983 |
AABR07067896.1
|
|
| chr3_-_45210474 | 22.77 |
ENSRNOT00000091777
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr4_+_29978739 | 21.46 |
ENSRNOT00000011756
|
Ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr13_-_80703615 | 20.27 |
ENSRNOT00000004599
|
Fmo4
|
flavin containing monooxygenase 4 |
| chr16_-_32868680 | 20.00 |
ENSRNOT00000015974
ENSRNOT00000082392 |
Aadat
|
aminoadipate aminotransferase |
| chr4_-_17594598 | 19.93 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr3_-_52447622 | 18.57 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr4_+_88832178 | 17.55 |
ENSRNOT00000088983
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr8_-_14880644 | 17.08 |
ENSRNOT00000015977
|
Fat3
|
FAT atypical cadherin 3 |
| chr7_-_82687130 | 16.88 |
ENSRNOT00000006791
|
Tmem74
|
transmembrane protein 74 |
| chr7_-_136853957 | 16.36 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr15_+_5306459 | 15.98 |
ENSRNOT00000092079
|
LOC108348081
|
CD99 antigen-like protein 2 |
| chr7_+_140788987 | 15.59 |
ENSRNOT00000086611
|
Kcnh3
|
potassium voltage-gated channel subfamily H member 3 |
| chr9_-_69953182 | 15.40 |
ENSRNOT00000015852
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr9_-_83253458 | 15.04 |
ENSRNOT00000041689
|
Epha4
|
Eph receptor A4 |
| chr7_+_11356118 | 14.86 |
ENSRNOT00000041325
|
Atcay
|
ATCAY, caytaxin |
| chr19_-_49510901 | 14.54 |
ENSRNOT00000082929
ENSRNOT00000079969 |
LOC687399
|
hypothetical protein LOC687399 |
| chr19_+_22450030 | 13.62 |
ENSRNOT00000021739
|
Neto2
|
neuropilin and tolloid like 2 |
| chr14_-_10395047 | 13.48 |
ENSRNOT00000002936
|
Gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr18_+_45023932 | 13.22 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr1_+_213676954 | 13.07 |
ENSRNOT00000050551
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chrX_+_57866709 | 13.07 |
ENSRNOT00000040923
|
Ppp4r3c
|
protein phosphatase 4 regulatory subunit 3C |
| chr10_-_47997796 | 13.05 |
ENSRNOT00000078422
|
Slc5a10
|
solute carrier family 5 member 10 |
| chr15_+_675085 | 12.99 |
ENSRNOT00000091318
|
Kcnma1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr1_+_15620653 | 12.46 |
ENSRNOT00000017401
|
Map7
|
microtubule-associated protein 7 |
| chrX_+_20351486 | 12.33 |
ENSRNOT00000093675
ENSRNOT00000047444 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr4_+_5644260 | 11.92 |
ENSRNOT00000041876
|
Actr3b
|
ARP3 actin related protein 3 homolog B |
| chr1_+_53531947 | 11.84 |
ENSRNOT00000077340
|
Tcp10b
|
t-complex protein 10b |
| chr2_-_199971965 | 11.63 |
ENSRNOT00000087026
|
Pde4dip
|
phosphodiesterase 4D interacting protein |
| chr16_+_74810938 | 11.59 |
ENSRNOT00000058091
|
Nek5
|
NIMA-related kinase 5 |
| chr7_+_120140460 | 11.57 |
ENSRNOT00000040513
ENSRNOT00000073905 |
Pdxp
|
pyridoxal phosphatase |
| chr15_-_45376476 | 11.27 |
ENSRNOT00000065838
|
Dleu7
|
deleted in lymphocytic leukemia, 7 |
| chr1_-_169344306 | 11.13 |
ENSRNOT00000022852
|
Ubqlnl
|
ubiquilin-like |
| chr11_+_61531571 | 10.95 |
ENSRNOT00000093467
ENSRNOT00000002727 |
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr1_+_97712177 | 10.60 |
ENSRNOT00000027745
|
MGC114499
|
similar to RIKEN cDNA 4930433I11 gene |
| chr3_+_69549673 | 10.58 |
ENSRNOT00000043974
|
Zfp804a
|
zinc finger protein 804A |
| chrX_-_1704033 | 10.58 |
ENSRNOT00000051956
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr1_-_250727079 | 10.56 |
ENSRNOT00000079942
ENSRNOT00000087588 |
Sgms1
|
sphingomyelin synthase 1 |
| chr4_+_114876770 | 10.52 |
ENSRNOT00000068102
ENSRNOT00000078175 |
Dctn1
|
dynactin subunit 1 |
| chr16_-_10726648 | 10.26 |
ENSRNOT00000080635
ENSRNOT00000087150 |
Sncg
|
synuclein, gamma |
| chr8_+_123126667 | 10.20 |
ENSRNOT00000015643
|
Osbpl10
|
oxysterol binding protein-like 10 |
| chr19_+_58505811 | 10.19 |
ENSRNOT00000031717
|
Map10
|
microtubule-associated protein 10 |
| chr13_+_71110064 | 10.12 |
ENSRNOT00000088329
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr5_-_171710312 | 10.08 |
ENSRNOT00000076044
ENSRNOT00000071280 ENSRNOT00000072059 ENSRNOT00000077015 |
Prdm16
|
PR/SET domain 16 |
| chr12_-_18526250 | 9.97 |
ENSRNOT00000001791
|
Asmt
|
acetylserotonin O-methyltransferase |
| chr9_-_65790347 | 9.79 |
ENSRNOT00000028506
|
AABR07067812.1
|
|
| chr2_-_192780631 | 9.70 |
ENSRNOT00000012371
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chrX_-_77295426 | 9.55 |
ENSRNOT00000090833
|
Taf9b
|
TATA-box binding protein associated factor 9b |
| chr16_+_49462889 | 9.51 |
ENSRNOT00000039909
|
Ankrd37
|
ankyrin repeat domain 37 |
| chrX_+_15035569 | 9.26 |
ENSRNOT00000006491
ENSRNOT00000078392 |
Porcn
|
porcupine homolog (Drosophila) |
| chr11_+_54137639 | 9.24 |
ENSRNOT00000066343
|
LOC100909977
|
leukocyte surface antigen CD47-like |
| chr10_-_18443934 | 9.11 |
ENSRNOT00000059895
ENSRNOT00000080021 |
Ranbp17
|
RAN binding protein 17 |
| chr1_-_222178725 | 9.00 |
ENSRNOT00000028697
|
Esrra
|
estrogen related receptor, alpha |
| chr4_+_114854458 | 9.00 |
ENSRNOT00000013312
|
LOC500227
|
hypothetical gene supported by BC079424 |
| chr1_+_187149453 | 8.98 |
ENSRNOT00000082738
|
Xylt1
|
xylosyltransferase 1 |
| chr5_+_120250616 | 8.92 |
ENSRNOT00000070967
|
Ak4
|
adenylate kinase 4 |
| chr1_-_227392054 | 8.84 |
ENSRNOT00000054806
|
Ms4a13-ps1
|
membrane-spanning 4-domains, subfamily A, member 13, pseudogene 1 |
| chr14_+_85871597 | 8.55 |
ENSRNOT00000079671
|
Ankrd36
|
ankyrin repeat domain 36 |
| chr1_-_99985422 | 8.55 |
ENSRNOT00000025701
|
Klk1c2
|
kallikrein 1-related peptidase C2 |
| chr11_+_42858478 | 8.53 |
ENSRNOT00000002293
|
Arl6
|
ADP-ribosylation factor like GTPase 6 |
| chr17_+_76306585 | 8.52 |
ENSRNOT00000065978
|
Dhtkd1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr3_-_161272460 | 8.49 |
ENSRNOT00000020740
|
Acot8
|
acyl-CoA thioesterase 8 |
| chr3_-_160922341 | 8.42 |
ENSRNOT00000029206
|
Tp53tg5
|
TP53 target 5 |
| chrY_+_914045 | 8.41 |
ENSRNOT00000088593
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr10_+_28449335 | 8.38 |
ENSRNOT00000029118
|
Atp10b
|
ATPase phospholipid transporting 10B (putative) |
| chr15_-_25505691 | 8.23 |
ENSRNOT00000074488
|
LOC100911492
|
homeobox protein OTX2-like |
| chr10_+_46216884 | 8.18 |
ENSRNOT00000004448
|
Nt5m
|
5',3'-nucleotidase, mitochondrial |
| chr1_+_79982639 | 8.13 |
ENSRNOT00000071916
|
Dmwd
|
dystrophia myotonica, WD repeat containing |
| chr3_+_56056925 | 8.10 |
ENSRNOT00000088351
ENSRNOT00000010508 |
Klhl23
|
kelch-like family member 23 |
| chr15_+_42960307 | 7.86 |
ENSRNOT00000012528
|
Trim35
|
tripartite motif-containing 35 |
| chrX_+_57866406 | 7.72 |
ENSRNOT00000076221
|
Ppp4r3c
|
protein phosphatase 4 regulatory subunit 3C |
| chr11_-_70211701 | 7.71 |
ENSRNOT00000076114
|
Muc13
|
mucin 13, cell surface associated |
| chr9_+_4807717 | 7.70 |
ENSRNOT00000092159
|
LOC100910526
|
sulfotransferase 1C2-like |
| chr16_-_10296341 | 7.62 |
ENSRNOT00000083990
|
Zfp488
|
zinc finger protein 488 |
| chr10_+_11146359 | 7.61 |
ENSRNOT00000006314
|
Pam16
|
presequence translocase associated motor 16 homolog |
| chr4_+_146455332 | 7.59 |
ENSRNOT00000009775
|
Hrh1
|
histamine receptor H 1 |
| chr10_+_55169282 | 7.45 |
ENSRNOT00000005423
|
Ccdc42
|
coiled-coil domain containing 42 |
| chr9_-_13311924 | 7.43 |
ENSRNOT00000015584
|
Kif6
|
kinesin family member 6 |
| chr7_-_12519154 | 7.26 |
ENSRNOT00000093376
ENSRNOT00000077681 |
Gpx4
|
glutathione peroxidase 4 |
| chr2_-_90568486 | 7.25 |
ENSRNOT00000059380
|
LOC100910852
|
uncharacterized LOC100910852 |
| chr14_-_78377825 | 7.09 |
ENSRNOT00000068104
|
AABR07015812.1
|
|
| chr2_-_89498395 | 6.98 |
ENSRNOT00000068507
|
AABR07009224.1
|
|
| chr6_+_43001948 | 6.94 |
ENSRNOT00000007374
|
Hpcal1
|
hippocalcin-like 1 |
| chr13_-_85622314 | 6.91 |
ENSRNOT00000005719
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr2_-_111793326 | 6.82 |
ENSRNOT00000092660
|
Nlgn1
|
neuroligin 1 |
| chr16_-_70998575 | 6.82 |
ENSRNOT00000019935
|
Kcnu1
|
potassium calcium-activated channel subfamily U member 1 |
| chr8_-_115274165 | 6.78 |
ENSRNOT00000056386
|
LOC102550160
|
IQ domain-containing protein F5-like |
| chr8_+_19994668 | 6.72 |
ENSRNOT00000050608
|
Olr1160
|
olfactory receptor 1160 |
| chr2_+_40554146 | 6.67 |
ENSRNOT00000015138
|
Pde4d
|
phosphodiesterase 4D |
| chr5_+_57947716 | 6.52 |
ENSRNOT00000067657
|
Dnai1
|
dynein, axonemal, intermediate chain 1 |
| chr12_-_6078411 | 6.30 |
ENSRNOT00000001197
|
Rxfp2
|
relaxin/insulin-like family peptide receptor 2 |
| chr4_+_168396775 | 6.25 |
ENSRNOT00000008645
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr9_-_81565416 | 6.24 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr3_+_72990358 | 6.20 |
ENSRNOT00000091920
|
Olr444
|
olfactory receptor 444 |
| chr14_+_22597103 | 6.16 |
ENSRNOT00000048482
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr9_-_14906410 | 6.14 |
ENSRNOT00000061674
|
LOC680955
|
hypothetical protein LOC680955 |
| chr13_-_90589466 | 6.11 |
ENSRNOT00000070812
|
Pea15
|
phosphoprotein enriched in astrocytes 15 |
| chr6_-_76535517 | 6.11 |
ENSRNOT00000083857
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr10_-_14299167 | 6.11 |
ENSRNOT00000042066
|
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr15_-_9086282 | 6.04 |
ENSRNOT00000008989
|
Thrb
|
thyroid hormone receptor beta |
| chr1_+_190072420 | 5.98 |
ENSRNOT00000037026
|
Abca15
|
ATP-binding cassette, subfamily A (ABC1), member 15 |
| chr4_-_100783750 | 5.82 |
ENSRNOT00000078956
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr6_+_129538982 | 5.81 |
ENSRNOT00000083626
ENSRNOT00000082522 |
Ak7
|
adenylate kinase 7 |
| chr5_+_129257429 | 5.80 |
ENSRNOT00000072396
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr3_-_103745236 | 5.68 |
ENSRNOT00000006876
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr11_-_64968437 | 5.67 |
ENSRNOT00000059541
|
Cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr3_+_75906945 | 5.63 |
ENSRNOT00000047110
|
Olr586
|
olfactory receptor 586 |
| chr16_+_12174370 | 5.60 |
ENSRNOT00000072045
|
LOC102553538
|
ral guanine nucleotide dissociation stimulator-like |
| chr7_-_139394166 | 5.58 |
ENSRNOT00000082429
|
Vdr
|
vitamin D (1,25- dihydroxyvitamin D3) receptor |
| chr14_-_92495894 | 5.56 |
ENSRNOT00000064483
|
Cobl
|
cordon-bleu WH2 repeat protein |
| chr1_-_204805738 | 5.55 |
ENSRNOT00000086830
|
Fam53b
|
family with sequence similarity 53, member B |
| chr14_+_11988671 | 5.53 |
ENSRNOT00000003207
|
Rasgef1b
|
RasGEF domain family, member 1B |
| chr17_+_78817529 | 5.52 |
ENSRNOT00000021918
|
Meig1
|
meiosis/spermiogenesis associated 1 |
| chr1_-_227359809 | 5.32 |
ENSRNOT00000088080
|
1700025F22Rik
|
RIKEN cDNA 1700025F22 gene |
| chr3_+_138770574 | 5.31 |
ENSRNOT00000012510
ENSRNOT00000066986 |
Dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr6_-_142418779 | 5.25 |
ENSRNOT00000072280
ENSRNOT00000065808 |
AABR07065814.1
|
|
| chr4_-_140503412 | 5.21 |
ENSRNOT00000079266
|
AABR07061707.1
|
|
| chr3_+_67870111 | 5.17 |
ENSRNOT00000012628
|
Nup35
|
nucleoporin 35 |
| chr3_+_161272385 | 5.12 |
ENSRNOT00000021052
|
Zswim3
|
zinc finger, SWIM-type containing 3 |
| chr12_-_18303041 | 5.07 |
ENSRNOT00000032844
|
LOC288521
|
similar to Leukosialin precursor (Leucocyte sialoglycoprotein) (Sialophorin) (CD43) (W3/13 antigen) |
| chr6_+_44230985 | 5.03 |
ENSRNOT00000005858
|
Kidins220
|
kinase D-interacting substrate 220 |
| chr1_-_99964824 | 5.03 |
ENSRNOT00000046377
|
Klk1c6
|
kallikrein 1-related peptidase C6 |
| chr16_-_12194118 | 5.00 |
ENSRNOT00000071517
|
RGD1559804
|
similar to hypothetical protein 4930474N05 |
| chrX_-_65335987 | 5.00 |
ENSRNOT00000047128
|
AABR07038981.1
|
|
| chr5_-_156734541 | 4.93 |
ENSRNOT00000021036
|
LOC100909857
|
cytidine deaminase-like |
| chr1_-_219144610 | 4.92 |
ENSRNOT00000023526
|
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr11_-_1437732 | 4.86 |
ENSRNOT00000037345
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr4_+_87608301 | 4.77 |
ENSRNOT00000058702
|
Vom1r71
|
vomeronasal 1 receptor 71 |
| chr12_-_31629881 | 4.69 |
ENSRNOT00000001238
|
Piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr1_-_221251329 | 4.61 |
ENSRNOT00000031678
|
Tigd3
|
tigger transposable element derived 3 |
| chr18_+_59096643 | 4.60 |
ENSRNOT00000025325
|
Wdr7
|
WD repeat domain 7 |
| chr9_-_43276685 | 4.56 |
ENSRNOT00000022779
|
Actr1b
|
ARP1 actin-related protein 1 homolog B, centractin beta |
| chr17_-_32158538 | 4.52 |
ENSRNOT00000024141
|
Nqo2
|
NAD(P)H quinone dehydrogenase 2 |
| chr13_+_83721300 | 4.47 |
ENSRNOT00000082677
|
Adcy10
|
adenylate cyclase 10 (soluble) |
| chr5_-_102743417 | 4.46 |
ENSRNOT00000067389
|
Bnc2
|
basonuclin 2 |
| chr17_+_5281727 | 4.44 |
ENSRNOT00000024781
|
Isca1
|
iron-sulfur cluster assembly 1 |
| chr3_+_70327193 | 4.43 |
ENSRNOT00000089165
|
Fsip2
|
fibrous sheath-interacting protein 2 |
| chr1_+_229066045 | 4.40 |
ENSRNOT00000016454
|
Glyat
|
glycine-N-acyltransferase |
| chr2_-_80667481 | 4.37 |
ENSRNOT00000016784
|
Trio
|
trio Rho guanine nucleotide exchange factor |
| chr10_+_3227160 | 4.34 |
ENSRNOT00000088075
|
Ntan1
|
N-terminal asparagine amidase |
| chr13_-_47440682 | 4.23 |
ENSRNOT00000037679
ENSRNOT00000005729 ENSRNOT00000050354 ENSRNOT00000050859 |
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr3_+_168345152 | 4.19 |
ENSRNOT00000017654
|
Dok5
|
docking protein 5 |
| chr9_+_45901741 | 4.11 |
ENSRNOT00000059571
|
Npas2
|
neuronal PAS domain protein 2 |
| chr6_+_8669722 | 4.06 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr20_+_18833481 | 4.05 |
ENSRNOT00000080846
|
Bicc1
|
BicC family RNA binding protein 1 |
| chr18_+_68983545 | 4.04 |
ENSRNOT00000085317
|
Stard6
|
StAR-related lipid transfer domain containing 6 |
| chr3_+_141068023 | 4.04 |
ENSRNOT00000036527
|
Kiz
|
kizuna centrosomal protein |
| chr8_+_117246376 | 4.02 |
ENSRNOT00000074493
|
Ccdc71
|
coiled-coil domain containing 71 |
| chr8_-_83693472 | 3.97 |
ENSRNOT00000015947
|
Fam83b
|
family with sequence similarity 83, member B |
| chr15_+_39945095 | 3.91 |
ENSRNOT00000016826
|
Shisa2
|
shisa family member 2 |
| chr10_+_91830654 | 3.84 |
ENSRNOT00000005176
|
Wnt3
|
wingless-type MMTV integration site family, member 3 |
| chr1_+_86914137 | 3.84 |
ENSRNOT00000027006
|
Fbxo17
|
F-box protein 17 |
| chr1_+_171592797 | 3.74 |
ENSRNOT00000026607
|
Syt9
|
synaptotagmin 9 |
| chr1_-_169598835 | 3.74 |
ENSRNOT00000072996
|
Olr156
|
olfactory receptor 156 |
| chr1_-_215310013 | 3.73 |
ENSRNOT00000054858
|
AABR07006049.2
|
|
| chr6_+_78567970 | 3.72 |
ENSRNOT00000032743
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr7_-_62162453 | 3.71 |
ENSRNOT00000010720
|
Cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr16_-_12538117 | 3.67 |
ENSRNOT00000046081
|
AABR07024718.1
|
|
| chr7_-_144960527 | 3.65 |
ENSRNOT00000086554
|
Zfp385a
|
zinc finger protein 385A |
| chr1_+_220243473 | 3.62 |
ENSRNOT00000084650
ENSRNOT00000027091 |
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr19_+_24456976 | 3.52 |
ENSRNOT00000004900
|
Ucp1
|
uncoupling protein 1 |
| chr6_-_26051229 | 3.46 |
ENSRNOT00000005855
|
Bre
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr3_+_171213936 | 3.41 |
ENSRNOT00000031586
|
Pck1
|
phosphoenolpyruvate carboxykinase 1 |
| chr6_-_122239614 | 3.39 |
ENSRNOT00000005015
|
Galc
|
galactosylceramidase |
| chr5_+_59491096 | 3.38 |
ENSRNOT00000019737
ENSRNOT00000036366 ENSRNOT00000083068 |
Clta
|
clathrin, light chain A |
| chr8_+_41336340 | 3.36 |
ENSRNOT00000072049
|
Olr1225
|
olfactory receptor 1225 |
| chr2_+_86914989 | 3.34 |
ENSRNOT00000085164
ENSRNOT00000081966 |
LOC100360380
|
zinc finger protein 457-like |
| chr4_+_88184956 | 3.32 |
ENSRNOT00000077129
|
Vom1r83
|
vomeronasal 1 receptor 83 |
| chr14_-_21898284 | 3.25 |
ENSRNOT00000036314
|
Prr27
|
proline rich 27 |
| chr14_+_85230648 | 3.19 |
ENSRNOT00000089866
|
Ap1b1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr16_+_13050305 | 3.11 |
ENSRNOT00000077388
|
AABR07024735.1
|
|
| chr2_+_149899836 | 3.02 |
ENSRNOT00000086481
|
RGD1560324
|
similar to hypothetical protein C130079G13 |
| chr3_-_77883067 | 3.01 |
ENSRNOT00000087979
|
Olr677
|
olfactory receptor 677 |
| chrX_+_45637415 | 2.99 |
ENSRNOT00000050544
|
RGD1563378
|
similar to ferritin heavy polypeptide-like 17 |
| chr1_+_221236773 | 2.97 |
ENSRNOT00000051979
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr1_+_226091774 | 2.89 |
ENSRNOT00000027693
|
Fads3
|
fatty acid desaturase 3 |
| chr2_+_196304492 | 2.82 |
ENSRNOT00000028640
|
Lysmd1
|
LysM domain containing 1 |
| chr17_+_8558827 | 2.82 |
ENSRNOT00000016187
|
Il9
|
interleukin 9 |
| chr9_+_49647257 | 2.75 |
ENSRNOT00000021899
|
Mrps9
|
mitochondrial ribosomal protein S9 |
| chr7_-_12346475 | 2.69 |
ENSRNOT00000060708
|
Mum1
|
melanoma associated antigen (mutated) 1 |
| chr12_+_6403940 | 2.66 |
ENSRNOT00000083484
|
B3glct
|
beta 3-glucosyltransferase |
| chr1_-_238376841 | 2.59 |
ENSRNOT00000076393
|
Tmc1
|
transmembrane channel-like 1 |
| chr1_-_8878136 | 2.58 |
ENSRNOT00000064836
ENSRNOT00000075850 |
Vta1
|
vesicle trafficking 1 |
| chr1_+_41065716 | 2.57 |
ENSRNOT00000035962
|
Ccdc170
|
coiled-coil domain containing 170 |
| chr12_-_30566032 | 2.55 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr8_-_43796224 | 2.50 |
ENSRNOT00000084315
|
Olr1330
|
olfactory receptor 1330 |
| chr1_+_264741911 | 2.50 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr3_-_153454160 | 2.50 |
ENSRNOT00000010298
|
Ghrh
|
growth hormone releasing hormone |
| chr15_-_28446785 | 2.42 |
ENSRNOT00000015439
|
Olr1638
|
olfactory receptor 1638 |
| chr11_-_43992598 | 2.32 |
ENSRNOT00000002260
|
Cldnd1
|
claudin domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rxra
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 20.0 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 6.1 | 24.6 | GO:0051958 | methotrexate transport(GO:0051958) |
| 5.9 | 17.6 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 5.7 | 22.9 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 5.0 | 15.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 5.0 | 14.9 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 4.5 | 13.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 4.3 | 13.0 | GO:0060082 | eye blink reflex(GO:0060082) |
| 4.1 | 12.3 | GO:2000682 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 3.9 | 11.6 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 3.6 | 21.5 | GO:1900272 | negative regulation of long-term synaptic potentiation(GO:1900272) |
| 3.3 | 10.0 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 2.8 | 8.5 | GO:1903445 | protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 2.7 | 8.2 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 2.5 | 7.6 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 2.2 | 15.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 2.2 | 6.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 2.1 | 10.5 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 1.9 | 5.6 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 1.7 | 6.8 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 1.6 | 9.4 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 1.5 | 10.6 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 1.5 | 10.2 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 1.4 | 18.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.4 | 5.7 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 1.4 | 9.7 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 1.3 | 10.3 | GO:0071464 | cellular response to hydrostatic pressure(GO:0071464) |
| 1.2 | 3.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 1.1 | 10.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 1.1 | 5.5 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 1.0 | 12.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 1.0 | 6.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 1.0 | 8.8 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.9 | 3.7 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.9 | 3.4 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.8 | 10.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.8 | 8.5 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.7 | 9.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.7 | 13.1 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 0.7 | 18.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.7 | 6.8 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.7 | 4.7 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.6 | 4.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.6 | 3.0 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.6 | 12.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.6 | 8.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.6 | 1.7 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.6 | 3.4 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.6 | 13.9 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.6 | 23.2 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.5 | 4.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.5 | 19.9 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) semaphorin-plexin signaling pathway(GO:0071526) |
| 0.5 | 2.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.5 | 4.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.5 | 6.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.5 | 7.7 | GO:0051923 | sulfation(GO:0051923) |
| 0.5 | 8.9 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.5 | 26.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.5 | 7.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.5 | 1.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.4 | 2.2 | GO:1903181 | myoblast fate commitment(GO:0048625) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.4 | 42.0 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.4 | 2.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.4 | 2.5 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.4 | 2.1 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.4 | 7.9 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.4 | 6.1 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.4 | 6.0 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.4 | 5.0 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.4 | 3.9 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.4 | 3.5 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.3 | 4.2 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.3 | 2.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.3 | 10.9 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.3 | 7.6 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.3 | 8.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.3 | 2.1 | GO:0035338 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.3 | 8.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.3 | 4.9 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.3 | 5.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 5.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) nuclear pore organization(GO:0006999) |
| 0.3 | 26.5 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.3 | 9.0 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.3 | 16.2 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.3 | 4.5 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.3 | 9.5 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.3 | 2.8 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.2 | 3.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 6.3 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.2 | 5.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 8.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.2 | 6.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 4.5 | GO:0043586 | tongue development(GO:0043586) |
| 0.2 | 4.4 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.2 | 11.6 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.2 | 6.1 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.2 | 7.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.2 | 18.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 1.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 3.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 27.7 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.1 | 9.3 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.1 | 1.9 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 6.2 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.1 | 7.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 15.8 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.1 | 6.2 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 0.9 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.7 | GO:0001845 | phagolysosome assembly(GO:0001845) equilibrioception(GO:0050957) |
| 0.1 | 14.1 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 3.5 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 12.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 1.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.8 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 8.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 6.8 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 1.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 4.1 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 1.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.3 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 3.8 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.7 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 4.0 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 2.0 | GO:1990748 | cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 5.5 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 23.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.7 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 3.3 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 22.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 5.4 | 21.5 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 1.9 | 7.6 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 1.9 | 15.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 1.7 | 8.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 1.6 | 6.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 1.3 | 13.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.2 | 10.6 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 1.1 | 6.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 1.1 | 8.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.9 | 18.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.9 | 11.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.9 | 10.9 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.8 | 5.6 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.8 | 5.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.7 | 9.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.7 | 13.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.7 | 3.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.7 | 10.2 | GO:0097431 | mitotic spindle pole(GO:0097431) mitotic spindle midzone(GO:1990023) |
| 0.6 | 1.7 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.5 | 4.7 | GO:0097433 | dense body(GO:0097433) |
| 0.5 | 5.2 | GO:0044615 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.4 | 8.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.4 | 2.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.4 | 6.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 20.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.4 | 3.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.4 | 15.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 7.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.3 | 15.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 9.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 57.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 0.8 | GO:0031308 | intrinsic component of nuclear outer membrane(GO:0031308) integral component of nuclear outer membrane(GO:0031309) |
| 0.3 | 3.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 3.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 0.7 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 6.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 10.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 2.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 6.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 9.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 13.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 29.1 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 3.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.7 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 8.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 5.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 5.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 10.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 5.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 11.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 22.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 1.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 36.7 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 7.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 3.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 3.6 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.1 | 17.0 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 5.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 9.0 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 13.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 43.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 6.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 6.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 4.1 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 24.6 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 6.0 | 42.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 5.7 | 22.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 5.0 | 20.0 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 5.0 | 19.8 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 3.9 | 23.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 3.9 | 11.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 3.0 | 15.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 3.0 | 9.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 2.8 | 8.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 2.2 | 15.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 2.1 | 10.6 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 2.0 | 17.6 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 1.8 | 47.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 1.7 | 20.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 1.5 | 19.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.5 | 7.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 1.5 | 5.8 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 1.4 | 5.6 | GO:0038181 | bile acid receptor activity(GO:0038181) vitamin D response element binding(GO:0070644) |
| 1.3 | 13.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 1.3 | 7.8 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 1.2 | 9.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.2 | 13.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 1.2 | 4.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 1.1 | 9.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 1.0 | 12.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 1.0 | 8.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.9 | 5.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.9 | 7.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.9 | 3.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.9 | 6.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.8 | 2.5 | GO:0031770 | growth hormone-releasing hormone receptor binding(GO:0031770) |
| 0.8 | 19.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.7 | 4.4 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.7 | 8.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.7 | 6.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.6 | 4.5 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.6 | 17.7 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.6 | 5.7 | GO:0016531 | metallochaperone activity(GO:0016530) copper chaperone activity(GO:0016531) |
| 0.6 | 6.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.5 | 10.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.5 | 10.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.5 | 4.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.5 | 6.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.5 | 8.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.5 | 8.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.5 | 6.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.4 | 33.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.4 | 14.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.4 | 4.2 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.4 | 10.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.4 | 3.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.4 | 1.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.4 | 5.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.3 | 6.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 11.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 7.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 2.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 9.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.3 | 13.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.3 | 5.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.3 | 3.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 10.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 16.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 10.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 2.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 4.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 3.8 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 2.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.2 | 1.0 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 5.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 0.7 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.2 | 4.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 3.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 4.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 10.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 8.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 10.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 3.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 2.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 7.4 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.8 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 13.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 3.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 7.3 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 0.9 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 1.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 4.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 7.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 2.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 6.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 10.6 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 11.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 6.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 3.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 7.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 2.2 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 16.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 3.6 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 5.1 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 7.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 2.8 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 7.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.8 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 5.5 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 6.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 19.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.4 | 15.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.3 | 10.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 6.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 4.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 17.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 4.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 7.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 5.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 4.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 3.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 13.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 6.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 16.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 3.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 8.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.8 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 21.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 2.2 | 20.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 1.7 | 24.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.1 | 27.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.9 | 8.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.7 | 27.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.7 | 11.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.6 | 8.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.5 | 7.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 10.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.4 | 3.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 3.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 10.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.3 | 6.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 15.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 13.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 6.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 6.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 16.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 5.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 6.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 4.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 7.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 15.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.2 | 15.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.2 | 5.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.2 | 4.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 8.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 2.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 4.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 4.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 4.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.9 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 3.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |