Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Runx3
Z-value: 0.42
Transcription factors associated with Runx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx3
|
ENSRNOG00000054217 | runt-related transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx3 | rn6_v1_chr5_+_153507093_153507093 | 0.16 | 4.8e-03 | Click! |
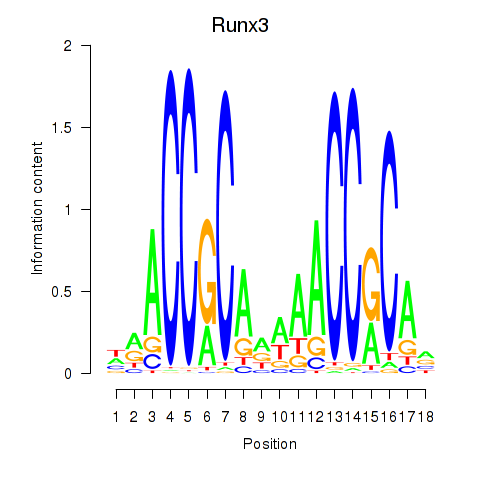
Activity profile of Runx3 motif
Sorted Z-values of Runx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_62948720 | 7.79 |
ENSRNOT00000075476
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr7_-_126913585 | 6.95 |
ENSRNOT00000036025
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr4_-_170763916 | 6.77 |
ENSRNOT00000071512
|
Hist1h4m
|
histone cluster 1, H4m |
| chr1_+_198528635 | 6.67 |
ENSRNOT00000022765
|
LOC308990
|
hypothetical protein LOC308990 |
| chr4_-_170092848 | 6.44 |
ENSRNOT00000072147
|
Hist2h4
|
histone cluster 2, H4 |
| chr2_+_225827504 | 6.41 |
ENSRNOT00000018343
|
Gclm
|
glutamate cysteine ligase, modifier subunit |
| chr12_-_22417980 | 6.07 |
ENSRNOT00000072208
|
Ephb4
|
EPH receptor B4 |
| chr4_-_170117325 | 5.14 |
ENSRNOT00000074198
|
LOC100912564
|
histone H4-like |
| chr4_-_170145262 | 5.02 |
ENSRNOT00000074526
|
Hist1h4b
|
histone cluster 1, H4b |
| chr17_-_44815995 | 4.90 |
ENSRNOT00000091201
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr17_+_43673294 | 4.55 |
ENSRNOT00000074109
|
Hist2h4a
|
histone cluster 2, H4 |
| chrX_+_68891227 | 4.45 |
ENSRNOT00000009635
|
Efnb1
|
ephrin B1 |
| chr5_+_105230580 | 4.40 |
ENSRNOT00000010056
|
Acer2
|
alkaline ceramidase 2 |
| chr5_+_149077412 | 4.38 |
ENSRNOT00000014666
|
Matn1
|
matrilin 1, cartilage matrix protein |
| chr9_-_61418679 | 4.13 |
ENSRNOT00000078800
|
Ankrd44
|
ankyrin repeat domain 44 |
| chr4_+_22898527 | 3.95 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr4_+_119680112 | 3.93 |
ENSRNOT00000013282
|
Gp9
|
glycoprotein IX (platelet) |
| chr1_+_224800252 | 3.91 |
ENSRNOT00000024488
|
Slc22a8
|
solute carrier family 22 member 8 |
| chr17_-_27665266 | 3.69 |
ENSRNOT00000060218
|
Rreb1
|
ras responsive element binding protein 1 |
| chr10_-_88144468 | 3.66 |
ENSRNOT00000084619
|
Ka11
|
type I keratin KA11 |
| chr13_+_89386023 | 3.65 |
ENSRNOT00000086223
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr1_+_214203861 | 3.31 |
ENSRNOT00000023219
|
Rassf7
|
Ras association domain family member 7 |
| chr19_+_43251539 | 3.29 |
ENSRNOT00000024793
|
Ddx19a
|
DEAD-box helicase 19A |
| chr4_+_49056010 | 3.18 |
ENSRNOT00000038566
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr7_-_120077612 | 3.16 |
ENSRNOT00000011750
|
Lgals2
|
galectin 2 |
| chr6_+_107245820 | 3.06 |
ENSRNOT00000012757
|
Papln
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr10_-_87928251 | 2.79 |
ENSRNOT00000064120
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr5_-_161981441 | 2.78 |
ENSRNOT00000020316
|
Pdpn
|
podoplanin |
| chr2_-_210934749 | 2.77 |
ENSRNOT00000026710
|
Gnai3
|
G protein subunit alpha i3 |
| chr3_-_13978224 | 2.64 |
ENSRNOT00000025528
|
Phf19
|
PHD finger protein 19 |
| chr15_+_47293699 | 2.52 |
ENSRNOT00000016503
|
Sox7
|
SRY box 7 |
| chr13_+_92249759 | 2.32 |
ENSRNOT00000089352
|
Olr1596
|
olfactory receptor 1596 |
| chr10_-_88144625 | 2.21 |
ENSRNOT00000086209
|
Ka11
|
type I keratin KA11 |
| chr1_-_72468738 | 2.11 |
ENSRNOT00000029136
|
Zfp628
|
zinc finger protein 628 |
| chr10_-_110701137 | 2.09 |
ENSRNOT00000074193
|
Znf750
|
zinc finger protein 750 |
| chr10_+_59533480 | 2.04 |
ENSRNOT00000087723
|
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr13_-_92988137 | 2.01 |
ENSRNOT00000004803
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chr3_+_45683993 | 1.93 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr1_-_142114634 | 1.85 |
ENSRNOT00000029471
|
Rccd1
|
RCC1 domain containing 1 |
| chrX_+_78259409 | 1.84 |
ENSRNOT00000049779
|
RGD1560455
|
similar to RIKEN cDNA A630033H20 gene |
| chr17_+_53956203 | 1.83 |
ENSRNOT00000022483
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr5_+_18963937 | 1.78 |
ENSRNOT00000082188
|
AABR07047089.1
|
|
| chr2_+_210034095 | 1.71 |
ENSRNOT00000024473
|
Lamtor5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr20_-_5244386 | 1.57 |
ENSRNOT00000070886
|
RT1-DMa
|
RT1 class II, locus DMa |
| chr7_+_132857628 | 1.39 |
ENSRNOT00000005438
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr4_+_147832136 | 1.33 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr14_-_86117578 | 1.31 |
ENSRNOT00000019288
|
Pold2
|
DNA polymerase delta 2, accessory subunit |
| chr2_-_181900856 | 1.28 |
ENSRNOT00000082156
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr8_-_7426611 | 1.26 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr4_+_100209951 | 1.25 |
ENSRNOT00000015807
|
LOC691113
|
hypothetical protein LOC691113 |
| chr1_-_221087449 | 1.18 |
ENSRNOT00000016987
|
Fam89b
|
family with sequence similarity 89, member B |
| chr6_-_145777767 | 1.06 |
ENSRNOT00000046799
|
Rpl32
|
ribosomal protein L32 |
| chr4_+_6827429 | 1.06 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr8_-_116872215 | 1.02 |
ENSRNOT00000045410
|
Apeh
|
acylaminoacyl-peptide hydrolase |
| chr2_-_193063357 | 0.97 |
ENSRNOT00000072995
|
AABR07012317.2
|
|
| chr20_-_3386048 | 0.96 |
ENSRNOT00000089550
ENSRNOT00000065025 |
Dhx16
|
DEAH-box helicase 16 |
| chr14_+_108412152 | 0.95 |
ENSRNOT00000083707
|
Pus10
|
pseudouridylate synthase 10 |
| chr2_-_198120041 | 0.94 |
ENSRNOT00000079621
ENSRNOT00000028747 |
Plekho1
|
pleckstrin homology domain containing O1 |
| chr16_-_69195097 | 0.90 |
ENSRNOT00000018973
|
Erlin2
|
ER lipid raft associated 2 |
| chr1_+_69789110 | 0.90 |
ENSRNOT00000088123
|
NEWGENE_1305281
|
hippocampus abundant transcript 1 |
| chr5_-_151898022 | 0.90 |
ENSRNOT00000000133
|
Pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr5_-_2882668 | 0.88 |
ENSRNOT00000049741
|
Terf1
|
telomeric repeat binding factor 1 |
| chr10_+_63677396 | 0.87 |
ENSRNOT00000005100
|
Slc43a2
|
solute carrier family 43 member 2 |
| chr4_+_115473811 | 0.87 |
ENSRNOT00000042875
|
Nagk
|
N-acetylglucosamine kinase |
| chr9_-_43139870 | 0.85 |
ENSRNOT00000021956
|
Sema4c
|
semaphorin 4C |
| chr6_+_108831108 | 0.83 |
ENSRNOT00000038594
|
LOC103692678
|
U6 snRNA-associated Sm-like protein LSm5 pseudogene |
| chr2_-_9023104 | 0.82 |
ENSRNOT00000039652
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr7_+_14589520 | 0.80 |
ENSRNOT00000060210
|
Cyp4f37
|
cytochrome P450, family 4, subfamily f, polypeptide 37 |
| chr1_-_77830399 | 0.69 |
ENSRNOT00000052231
|
LOC100360449
|
ribosomal protein L9-like |
| chr10_+_11046221 | 0.67 |
ENSRNOT00000005109
|
Nmral1
|
NmrA like redox sensor 1 |
| chr15_+_30812576 | 0.65 |
ENSRNOT00000060420
|
AABR07017768.1
|
|
| chr20_+_4966817 | 0.64 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_116372519 | 0.61 |
ENSRNOT00000031730
|
Lrrc34
|
leucine rich repeat containing 34 |
| chr15_-_29976615 | 0.60 |
ENSRNOT00000060396
|
AABR07017676.1
|
|
| chr10_-_15667955 | 0.58 |
ENSRNOT00000085746
ENSRNOT00000027940 |
Mpg
|
N-methylpurine-DNA glycosylase |
| chr14_+_78939903 | 0.58 |
ENSRNOT00000078562
ENSRNOT00000088221 |
Man2b2
Mrfap1
|
mannosidase, alpha, class 2B, member 2 Morf4 family associated protein 1 |
| chr10_-_61744976 | 0.58 |
ENSRNOT00000079926
ENSRNOT00000092314 ENSRNOT00000034298 |
Sgsm2
|
small G protein signaling modulator 2 |
| chr20_+_4967194 | 0.52 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr3_-_10161989 | 0.51 |
ENSRNOT00000012312
|
Exosc2
|
exosome component 2 |
| chr1_+_69788609 | 0.45 |
ENSRNOT00000075340
|
NEWGENE_1305281
|
hippocampus abundant transcript 1 |
| chrX_+_157353577 | 0.42 |
ENSRNOT00000090596
|
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr10_+_11046024 | 0.39 |
ENSRNOT00000084850
|
Nmral1
|
NmrA like redox sensor 1 |
| chr13_+_110372164 | 0.38 |
ENSRNOT00000049634
|
RGD1560186
|
similar to ribosomal protein L37 |
| chr7_+_2504695 | 0.37 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr7_+_2827247 | 0.35 |
ENSRNOT00000090689
|
Rnf41
|
ring finger protein 41 |
| chr6_-_115352681 | 0.34 |
ENSRNOT00000005873
|
Gtf2a1
|
general transcription factor 2A subunit 1 |
| chr5_+_57738321 | 0.33 |
ENSRNOT00000063849
|
Ubap1
|
ubiquitin-associated protein 1 |
| chr10_+_48240330 | 0.30 |
ENSRNOT00000057798
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr5_-_25721072 | 0.30 |
ENSRNOT00000021839
|
Tmem67
|
transmembrane protein 67 |
| chr2_-_219693629 | 0.26 |
ENSRNOT00000020907
|
Mfsd14a
|
major facilitator superfamily domain containing 14A |
| chr9_+_10995198 | 0.21 |
ENSRNOT00000089071
|
Ubxn6
|
UBX domain protein 6 |
| chr10_-_85435016 | 0.21 |
ENSRNOT00000079921
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr12_+_2228670 | 0.19 |
ENSRNOT00000001327
|
Trappc5
|
trafficking protein particle complex 5 |
| chr10_-_14373334 | 0.12 |
ENSRNOT00000022050
|
Cramp1
|
cramped chromatin regulator homolog 1 |
| chrX_+_63343546 | 0.11 |
ENSRNOT00000076315
|
Klhl15
|
kelch-like family member 15 |
| chr1_-_235813030 | 0.05 |
ENSRNOT00000014800
|
Vps13a
|
vacuolar protein sorting 13A |
| chr10_+_48240127 | 0.03 |
ENSRNOT00000080682
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 1.9 | 32.8 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 1.6 | 6.4 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 1.5 | 6.1 | GO:2000523 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 1.5 | 4.4 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 1.3 | 3.9 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.8 | 3.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.7 | 2.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.6 | 4.4 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.6 | 1.8 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.5 | 1.4 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) regulation of peroxidase activity(GO:2000468) |
| 0.4 | 1.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.4 | 1.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.3 | 2.8 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.3 | 2.6 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.3 | 0.9 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.3 | 1.6 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.2 | 3.6 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.2 | 0.9 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 4.4 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.2 | 0.6 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.2 | 0.9 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 0.5 | GO:0071031 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.2 | 1.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.2 | 1.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 0.9 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 1.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 0.8 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 1.9 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 2.8 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.1 | 1.3 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 0.9 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 1.7 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 1.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 3.3 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.0 | 2.5 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.8 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.3 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 1.0 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.3 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) photoreceptor cell outer segment organization(GO:0035845) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.9 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 1.1 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 3.3 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 3.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 32.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 1.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.3 | 1.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 1.7 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 1.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 0.9 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.2 | 0.9 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 2.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.8 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.1 | 2.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 4.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.9 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 2.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 1.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 3.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 4.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 3.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 5.3 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 3.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.7 | 3.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.5 | 3.9 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.5 | 1.4 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.4 | 2.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.4 | 3.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 1.8 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.3 | 2.8 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.3 | 6.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.3 | 6.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.3 | 0.9 | GO:0009384 | N-acylmannosamine kinase activity(GO:0009384) |
| 0.2 | 2.0 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 34.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.3 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 0.9 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 4.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.0 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 2.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 1.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 4.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 3.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.6 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.9 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 3.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 0.9 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0017022 | myosin binding(GO:0017022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 3.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 2.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 3.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 4.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 4.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.4 | 3.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 3.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 6.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 1.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 2.8 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 4.0 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 4.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |