Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
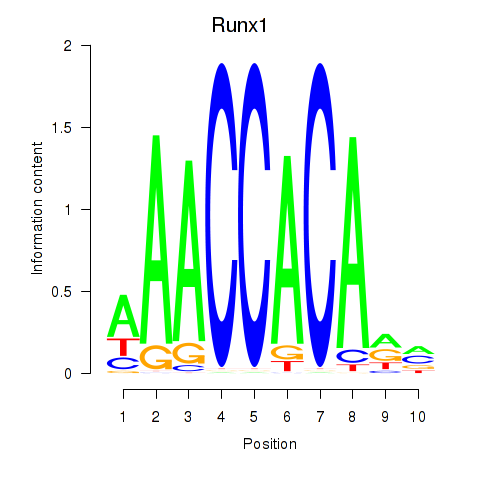
Results for Runx1
Z-value: 2.07
Transcription factors associated with Runx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx1
|
ENSRNOG00000001704 | runt-related transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx1 | rn6_v1_chr11_-_33003021_33003021 | 0.90 | 5.4e-114 | Click! |
Activity profile of Runx1 motif
Sorted Z-values of Runx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_206342066 | 141.47 |
ENSRNOT00000026556
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr1_-_226887156 | 112.68 |
ENSRNOT00000054809
ENSRNOT00000028347 |
Cd6
|
Cd6 molecule |
| chr3_-_44177689 | 112.51 |
ENSRNOT00000006387
|
Cytip
|
cytohesin 1 interacting protein |
| chr11_-_33003021 | 107.82 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr10_-_31493419 | 98.44 |
ENSRNOT00000009211
|
Itk
|
IL2-inducible T-cell kinase |
| chr14_+_3058993 | 94.34 |
ENSRNOT00000002807
|
Gfi1
|
growth factor independent 1 transcriptional repressor |
| chr10_-_56506446 | 83.00 |
ENSRNOT00000021357
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr9_+_18564927 | 75.84 |
ENSRNOT00000061014
|
Runx2
|
runt-related transcription factor 2 |
| chr11_-_60547201 | 74.56 |
ENSRNOT00000093151
|
Btla
|
B and T lymphocyte associated |
| chr1_-_198577226 | 73.60 |
ENSRNOT00000055013
|
Spn
|
sialophorin |
| chrX_+_15155230 | 73.46 |
ENSRNOT00000073289
ENSRNOT00000051439 |
Was
|
Wiskott-Aldrich syndrome |
| chr2_-_149325913 | 72.11 |
ENSRNOT00000036690
|
Gpr171
|
G protein-coupled receptor 171 |
| chr10_-_70871066 | 71.03 |
ENSRNOT00000015139
|
Ccl3
|
C-C motif chemokine ligand 3 |
| chr6_+_109562587 | 67.81 |
ENSRNOT00000011563
|
Batf
|
basic leucine zipper ATF-like transcription factor |
| chr6_-_137664133 | 64.40 |
ENSRNOT00000018613
|
Gpr132
|
G protein-coupled receptor 132 |
| chr11_-_60546997 | 62.96 |
ENSRNOT00000083124
ENSRNOT00000050092 |
Btla
|
B and T lymphocyte associated |
| chr2_-_203413124 | 61.63 |
ENSRNOT00000077898
ENSRNOT00000056140 |
Cd101
|
CD101 molecule |
| chr3_+_18970574 | 60.11 |
ENSRNOT00000088394
|
AABR07051665.1
|
|
| chr9_+_67546408 | 58.15 |
ENSRNOT00000013701
|
Cd28
|
Cd28 molecule |
| chr13_+_89774764 | 55.75 |
ENSRNOT00000005619
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr2_-_44981458 | 53.53 |
ENSRNOT00000014134
|
Gzma
|
granzyme A |
| chrX_+_14578264 | 52.85 |
ENSRNOT00000038994
|
Cybb
|
cytochrome b-245 beta chain |
| chr3_+_19141133 | 51.46 |
ENSRNOT00000058323
|
AABR07051675.1
|
|
| chr3_-_146491837 | 51.31 |
ENSRNOT00000061469
|
Vsx1
|
visual system homeobox 1 |
| chr3_+_19441604 | 46.37 |
ENSRNOT00000090581
|
AABR07051692.1
|
|
| chr13_+_92146586 | 44.47 |
ENSRNOT00000004659
|
Olr1588
|
olfactory receptor 1588 |
| chr20_-_2678141 | 43.48 |
ENSRNOT00000072377
ENSRNOT00000083833 |
C4a
|
complement component 4A (Rodgers blood group) |
| chr7_-_142352382 | 42.99 |
ENSRNOT00000048488
|
Galnt6
|
polypeptide N-acetylgalactosaminyltransferase 6 |
| chr20_-_4508197 | 42.78 |
ENSRNOT00000086027
ENSRNOT00000000514 |
C4a
|
complement component 4A (Rodgers blood group) |
| chr20_-_4316715 | 42.22 |
ENSRNOT00000031704
|
C4b
|
complement C4B (Chido blood group) |
| chr4_+_145413230 | 41.46 |
ENSRNOT00000056508
|
Il17re
|
interleukin 17 receptor E |
| chr7_-_144880092 | 40.60 |
ENSRNOT00000055281
|
Nfe2
|
nuclear factor, erythroid 2 |
| chr5_+_151362019 | 39.65 |
ENSRNOT00000077083
ENSRNOT00000067801 |
Wasf2
|
WAS protein family, member 2 |
| chrX_-_159841072 | 39.62 |
ENSRNOT00000001158
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr1_+_192233910 | 39.56 |
ENSRNOT00000016418
ENSRNOT00000016442 |
Prkcb
|
protein kinase C, beta |
| chr3_+_19366370 | 35.12 |
ENSRNOT00000086557
|
AABR07051689.1
|
|
| chr1_-_82004538 | 34.95 |
ENSRNOT00000087572
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr13_-_83202864 | 34.92 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr15_-_42947656 | 34.43 |
ENSRNOT00000030007
|
Ptk2b
|
protein tyrosine kinase 2 beta |
| chr13_-_92210052 | 31.70 |
ENSRNOT00000004670
|
Olr1592
|
olfactory receptor 1592 |
| chr10_-_88036040 | 31.51 |
ENSRNOT00000018851
|
Krt13
|
keratin 13 |
| chr3_+_18787606 | 30.78 |
ENSRNOT00000090508
|
AABR07051658.1
|
|
| chr1_-_82003691 | 29.31 |
ENSRNOT00000084569
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr1_+_228690119 | 28.69 |
ENSRNOT00000087169
|
Pfpl
|
pore forming protein-like |
| chr10_-_66873948 | 25.33 |
ENSRNOT00000039261
|
Evi2a
|
ecotropic viral integration site 2A |
| chr10_-_83655182 | 25.09 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr8_-_96132634 | 22.67 |
ENSRNOT00000041655
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr9_+_47165342 | 22.01 |
ENSRNOT00000020144
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr3_+_55369214 | 21.42 |
ENSRNOT00000067161
|
Nostrin
|
nitric oxide synthase trafficking |
| chr8_+_48569328 | 21.39 |
ENSRNOT00000084030
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr1_+_129255396 | 21.30 |
ENSRNOT00000040147
|
Fam169b
|
family with sequence similarity 169, member B |
| chr5_+_25253010 | 21.22 |
ENSRNOT00000061333
|
Rbm12b
|
RNA binding motif protein 12B |
| chr1_-_219544328 | 21.13 |
ENSRNOT00000025847
|
Grk2
|
G protein-coupled receptor kinase 2 |
| chr13_+_92249759 | 20.84 |
ENSRNOT00000089352
|
Olr1596
|
olfactory receptor 1596 |
| chr14_+_35683442 | 20.68 |
ENSRNOT00000003085
|
Chic2
|
cysteine-rich hydrophobic domain 2 |
| chr6_+_52631092 | 20.46 |
ENSRNOT00000014054
|
Atxn7l1
|
ataxin 7-like 1 |
| chr20_+_4852671 | 19.85 |
ENSRNOT00000001111
|
Lta
|
lymphotoxin alpha |
| chr5_+_25725683 | 19.73 |
ENSRNOT00000087602
|
LOC679087
|
similar to swan |
| chr8_-_40137390 | 19.37 |
ENSRNOT00000042717
|
Panx3
|
pannexin 3 |
| chr17_-_53688966 | 19.14 |
ENSRNOT00000078643
|
LOC100912163
|
AT-rich interactive domain-containing protein 4B-like |
| chr1_+_197999037 | 19.03 |
ENSRNOT00000091065
|
Apobr
|
apolipoprotein B receptor |
| chr16_-_6675746 | 18.88 |
ENSRNOT00000025858
|
Prkcd
|
protein kinase C, delta |
| chr20_+_2088533 | 18.36 |
ENSRNOT00000001012
ENSRNOT00000079021 |
Znrd1
|
zinc ribbon domain containing 1 |
| chr10_+_75087892 | 18.11 |
ENSRNOT00000065910
|
Mpo
|
myeloperoxidase |
| chr3_+_18805220 | 17.79 |
ENSRNOT00000071216
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr20_+_4852496 | 17.18 |
ENSRNOT00000088936
|
Lta
|
lymphotoxin alpha |
| chrX_-_15768522 | 16.34 |
ENSRNOT00000077391
|
Foxp3
|
forkhead box P3 |
| chr20_-_22459025 | 16.32 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr3_+_21764377 | 16.29 |
ENSRNOT00000063938
|
Gpr21
|
G protein-coupled receptor 21 |
| chr8_-_96266342 | 15.81 |
ENSRNOT00000078891
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr3_+_66673071 | 14.67 |
ENSRNOT00000034769
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr7_+_61337381 | 14.50 |
ENSRNOT00000009919
|
Ifng
|
interferon gamma |
| chr14_+_86828355 | 14.45 |
ENSRNOT00000088611
|
Ccm2
|
CCM2 scaffolding protein |
| chr11_+_88122271 | 12.00 |
ENSRNOT00000002540
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr1_+_197999336 | 11.87 |
ENSRNOT00000023555
|
Apobr
|
apolipoprotein B receptor |
| chr19_+_10024947 | 11.56 |
ENSRNOT00000061392
|
Cfap20
|
cilia and flagella associated protein 20 |
| chr9_+_71247781 | 11.21 |
ENSRNOT00000049654
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr5_-_86554102 | 11.13 |
ENSRNOT00000083340
ENSRNOT00000007710 |
Cdk5rap2
|
CDK5 regulatory subunit associated protein 2 |
| chr4_-_180722358 | 10.87 |
ENSRNOT00000040645
|
Itpr2
|
inositol 1,4,5-trisphosphate receptor, type 2 |
| chr9_-_15700235 | 10.25 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr14_-_81399353 | 10.15 |
ENSRNOT00000018340
|
Add1
|
adducin 1 |
| chr10_-_39604070 | 10.02 |
ENSRNOT00000032333
|
Csf2
|
colony stimulating factor 2 |
| chr2_+_116032455 | 10.01 |
ENSRNOT00000066495
|
Phc3
|
polyhomeotic homolog 3 |
| chr7_+_121930615 | 9.92 |
ENSRNOT00000091270
ENSRNOT00000033975 |
Tnrc6b
|
trinucleotide repeat containing 6B |
| chr10_+_104582955 | 8.55 |
ENSRNOT00000009733
|
Unk
|
unkempt family zinc finger |
| chr3_+_170399302 | 8.03 |
ENSRNOT00000035245
|
Cass4
|
Cas scaffolding protein family member 4 |
| chr3_+_66673246 | 7.86 |
ENSRNOT00000081338
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr10_+_64336200 | 7.62 |
ENSRNOT00000046519
|
Rpl37
|
ribosomal protein L37 |
| chr10_+_63677396 | 7.51 |
ENSRNOT00000005100
|
Slc43a2
|
solute carrier family 43 member 2 |
| chr16_+_83824430 | 7.19 |
ENSRNOT00000032918
|
Irs2
|
insulin receptor substrate 2 |
| chr9_+_95202632 | 7.13 |
ENSRNOT00000025652
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr2_+_205553163 | 6.56 |
ENSRNOT00000039572
|
Nras
|
neuroblastoma RAS viral oncogene homolog |
| chr1_+_100577056 | 6.20 |
ENSRNOT00000026992
|
Napsa
|
napsin A aspartic peptidase |
| chr8_-_48736506 | 5.50 |
ENSRNOT00000016227
|
Ccdc84
|
coiled-coil domain containing 84 |
| chr9_+_42871950 | 5.19 |
ENSRNOT00000089673
|
Arid5a
|
AT-rich interaction domain 5A |
| chr7_-_60778136 | 5.18 |
ENSRNOT00000029493
|
Slc35e3
|
solute carrier family 35, member E3 |
| chr7_-_62162453 | 5.17 |
ENSRNOT00000010720
|
Cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr1_+_277261681 | 4.88 |
ENSRNOT00000022724
|
Plekhs1
|
pleckstrin homology domain containing S1 |
| chr4_-_40036563 | 4.57 |
ENSRNOT00000084531
|
Tmem168
|
transmembrane protein 168 |
| chr14_-_84820415 | 4.37 |
ENSRNOT00000057501
|
Mtmr3
|
myotubularin related protein 3 |
| chr10_-_65200109 | 4.29 |
ENSRNOT00000030501
|
Nufip2
|
NUFIP2, FMR1 interacting protein 2 |
| chr15_-_27716008 | 3.67 |
ENSRNOT00000011741
|
Ttc5
|
tetratricopeptide repeat domain 5 |
| chr5_-_56753832 | 3.53 |
ENSRNOT00000073045
|
Sec61gl
|
SEC61 gamma subunit-like |
| chr1_+_192933540 | 3.37 |
ENSRNOT00000035770
|
Rbbp6
|
RB binding protein 6, ubiquitin ligase |
| chr9_-_81864202 | 3.29 |
ENSRNOT00000084964
|
Zfp142
|
zinc finger protein 142 |
| chr7_-_144322240 | 3.11 |
ENSRNOT00000089290
|
LOC100912282
|
calcium-binding and coiled-coil domain-containing protein 1-like |
| chr2_-_210550490 | 2.62 |
ENSRNOT00000081835
ENSRNOT00000025222 ENSRNOT00000086403 |
Csf1
|
colony stimulating factor 1 |
| chr3_+_104816987 | 2.35 |
ENSRNOT00000042103
ENSRNOT00000044625 |
Fmn1
|
formin 1 |
| chr7_-_145450233 | 2.27 |
ENSRNOT00000092974
ENSRNOT00000021523 |
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr17_-_22143324 | 2.17 |
ENSRNOT00000019361
|
Edn1
|
endothelin 1 |
| chr2_+_24546536 | 1.13 |
ENSRNOT00000014036
|
Otp
|
orthopedia homeobox |
| chr8_+_122076759 | 0.59 |
ENSRNOT00000012545
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr2_-_187622373 | 0.58 |
ENSRNOT00000026396
|
Rhbg
|
Rh family B glycoprotein |
| chr5_-_151898022 | 0.45 |
ENSRNOT00000000133
|
Pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr14_+_33010300 | 0.08 |
ENSRNOT00000002809
|
Igfbp7
|
insulin-like growth factor binding protein 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 58.8 | 176.4 | GO:0071661 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 35.9 | 107.8 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 31.4 | 94.3 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 21.4 | 64.3 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 19.3 | 57.8 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 17.6 | 52.9 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 17.2 | 34.4 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 14.2 | 71.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 13.8 | 137.5 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 13.2 | 39.6 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 12.6 | 75.8 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 12.3 | 37.0 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 12.2 | 73.5 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 8.9 | 98.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 8.7 | 112.7 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 8.2 | 24.5 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 8.0 | 72.4 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 7.3 | 22.0 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 6.4 | 38.5 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 6.2 | 67.8 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 6.0 | 18.1 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 6.0 | 72.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 5.7 | 51.3 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 5.4 | 16.3 | GO:0045077 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 5.4 | 16.3 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 5.3 | 42.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 4.8 | 58.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 4.1 | 40.6 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 3.7 | 11.2 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 3.0 | 21.1 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 2.7 | 86.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 2.6 | 31.5 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 2.4 | 7.2 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 2.2 | 19.4 | GO:0034214 | protein hexamerization(GO:0034214) |
| 1.9 | 11.6 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 1.9 | 39.6 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 1.7 | 5.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 1.7 | 10.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.6 | 14.4 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 1.6 | 11.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 1.6 | 10.9 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 1.4 | 64.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 1.3 | 2.6 | GO:1904124 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 1.1 | 12.0 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 1.1 | 2.2 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 1.0 | 57.0 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.9 | 9.9 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.9 | 8.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.6 | 22.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.6 | 18.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.6 | 2.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.5 | 16.3 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.4 | 6.2 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.4 | 20.7 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.3 | 31.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.3 | 6.6 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.2 | 30.9 | GO:0006641 | triglyceride metabolic process(GO:0006641) |
| 0.2 | 1.9 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.6 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 7.5 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.2 | 4.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.2 | 43.0 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.2 | 5.2 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.2 | 92.3 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.2 | 1.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 73.7 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.1 | 41.5 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.1 | 25.1 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.1 | 0.5 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 3.4 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 21.4 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 5.4 | GO:0016055 | Wnt signaling pathway(GO:0016055) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 7.6 | GO:0006412 | translation(GO:0006412) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 37.6 | 112.7 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 5.3 | 52.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 4.9 | 73.6 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 4.8 | 38.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) GAIT complex(GO:0097452) |
| 4.4 | 39.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 3.7 | 11.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 2.0 | 34.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.9 | 42.2 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 1.6 | 58.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 1.1 | 18.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 1.1 | 10.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 1.1 | 12.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 1.1 | 239.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 1.0 | 94.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.9 | 107.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.9 | 2.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.7 | 11.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.7 | 10.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.6 | 19.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.6 | 73.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.6 | 0.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.6 | 6.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.5 | 2.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.5 | 39.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.5 | 153.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.4 | 4.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.4 | 64.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.4 | 89.2 | GO:0005938 | cell cortex(GO:0005938) |
| 0.4 | 40.6 | GO:0016605 | PML body(GO:0016605) |
| 0.3 | 21.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 20.7 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.3 | 91.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.2 | 18.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 86.3 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 49.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 11.6 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 77.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 0.5 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 7.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 120.3 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 3.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 4.4 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 4.6 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 2.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 12.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 5.6 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.7 | 71.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 22.5 | 112.7 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 19.8 | 39.6 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 7.3 | 22.0 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 6.3 | 18.9 | GO:0070976 | TIR domain binding(GO:0070976) |
| 6.2 | 30.9 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 6.0 | 42.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 6.0 | 72.1 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 5.9 | 52.9 | GO:0016175 | NAD(P)H oxidase activity(GO:0016174) superoxide-generating NADPH oxidase activity(GO:0016175) |
| 5.4 | 16.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 5.2 | 41.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 4.9 | 34.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 4.8 | 19.4 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 3.9 | 43.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 3.7 | 11.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 3.5 | 21.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 3.2 | 73.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 3.2 | 107.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 2.9 | 58.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 2.8 | 22.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 2.7 | 10.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 2.4 | 38.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 2.4 | 12.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 2.0 | 16.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 1.7 | 142.1 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 1.6 | 98.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 1.5 | 66.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 1.4 | 10.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 1.4 | 8.6 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 1.3 | 2.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.2 | 40.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 1.1 | 75.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 1.0 | 34.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 1.0 | 37.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.9 | 4.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.7 | 2.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.7 | 16.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.5 | 132.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.5 | 65.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.5 | 18.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.4 | 18.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.4 | 86.3 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.4 | 29.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.4 | 102.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.4 | 42.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.3 | 7.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.3 | 2.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 6.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 5.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 39.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 7.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 72.0 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.1 | 7.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 11.7 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 101.1 | GO:0060089 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
| 0.0 | 35.5 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 6.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 3.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 10.0 | GO:0008270 | zinc ion binding(GO:0008270) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 76.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 3.4 | 276.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 2.6 | 34.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 2.5 | 172.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 2.1 | 139.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 2.0 | 53.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 2.0 | 83.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 1.5 | 77.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 1.5 | 92.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 1.3 | 105.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 1.3 | 39.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 1.2 | 26.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 1.1 | 18.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.8 | 14.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.6 | 10.9 | ST ADRENERGIC | Adrenergic Pathway |
| 0.5 | 16.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 2.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 3.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 2.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 171.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 6.2 | 86.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 3.8 | 34.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 3.4 | 58.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 2.8 | 137.5 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 2.7 | 75.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 2.6 | 39.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 2.6 | 73.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 2.1 | 106.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 2.1 | 39.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 1.5 | 52.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 1.4 | 21.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 1.2 | 41.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 1.0 | 14.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.9 | 13.7 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.8 | 43.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.5 | 9.9 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.4 | 40.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.4 | 66.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.3 | 10.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 7.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 5.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 31.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 11.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 4.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 7.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 5.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |