Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Rora
Z-value: 0.56
Transcription factors associated with Rora
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rora
|
ENSRNOG00000027145 | RAR-related orphan receptor A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rora | rn6_v1_chr8_+_75516904_75516904 | 0.14 | 1.3e-02 | Click! |
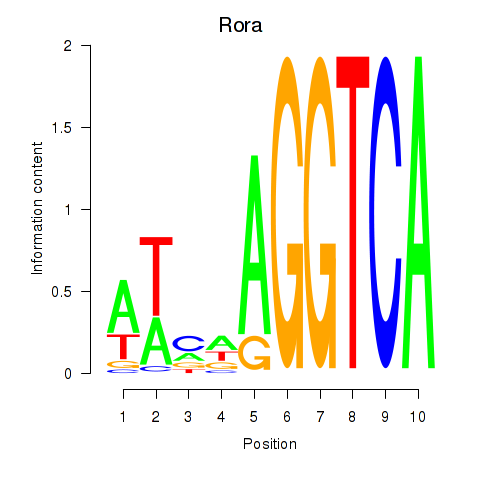
Activity profile of Rora motif
Sorted Z-values of Rora motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_101687855 | 19.87 |
ENSRNOT00000028546
ENSRNOT00000091597 |
Dbp
|
D-box binding PAR bZIP transcription factor |
| chr19_-_10653800 | 12.48 |
ENSRNOT00000022128
|
Cx3cl1
|
C-X3-C motif chemokine ligand 1 |
| chr3_-_113525864 | 10.53 |
ENSRNOT00000046659
|
Frmd5
|
FERM domain containing 5 |
| chr8_-_82533689 | 10.16 |
ENSRNOT00000014124
|
Tmod2
|
tropomodulin 2 |
| chr3_+_113319456 | 10.09 |
ENSRNOT00000051354
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr7_-_81592206 | 9.97 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr10_-_86688730 | 9.25 |
ENSRNOT00000055333
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr6_-_136145837 | 8.76 |
ENSRNOT00000015122
|
Ckb
|
creatine kinase B |
| chr18_-_1946840 | 8.48 |
ENSRNOT00000041878
|
Abhd3
|
abhydrolase domain containing 3 |
| chr13_-_86451002 | 7.91 |
ENSRNOT00000043004
ENSRNOT00000027996 |
Pbx1
|
PBX homeobox 1 |
| chr12_+_47024442 | 7.77 |
ENSRNOT00000001545
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chr14_-_114583122 | 7.69 |
ENSRNOT00000084595
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr5_-_147412705 | 7.50 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr8_-_47404010 | 7.38 |
ENSRNOT00000038647
|
Tmem136
|
transmembrane protein 136 |
| chr13_+_34610684 | 7.08 |
ENSRNOT00000093019
ENSRNOT00000003280 |
Tfcp2l1
|
transcription factor CP2-like 1 |
| chr9_+_61692154 | 7.04 |
ENSRNOT00000082300
|
Hspe1
|
heat shock protein family E member 1 |
| chr3_+_8534440 | 6.91 |
ENSRNOT00000045827
ENSRNOT00000082672 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr1_+_199555722 | 6.64 |
ENSRNOT00000054983
|
Itgax
|
integrin subunit alpha X |
| chr18_+_2416871 | 6.52 |
ENSRNOT00000081399
|
Gata6
|
GATA binding protein 6 |
| chr2_-_210282352 | 6.44 |
ENSRNOT00000075653
|
Slc6a17
|
solute carrier family 6 member 17 |
| chr3_+_151126591 | 6.35 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr18_+_2416552 | 6.30 |
ENSRNOT00000030726
|
Gata6
|
GATA binding protein 6 |
| chr19_+_568287 | 6.18 |
ENSRNOT00000016419
|
Cdh16
|
cadherin 16 |
| chr9_-_61690956 | 6.13 |
ENSRNOT00000066589
|
Hspd1
|
heat shock protein family D member 1 |
| chr10_+_55712043 | 6.12 |
ENSRNOT00000010141
|
Aloxe3
|
arachidonate lipoxygenase 3 |
| chr1_+_197659187 | 6.10 |
ENSRNOT00000082228
|
LOC103690016
|
serine/threonine-protein kinase SBK1 |
| chr5_-_157268903 | 6.09 |
ENSRNOT00000022716
|
Pla2g5
|
phospholipase A2, group V |
| chr7_+_53630621 | 5.99 |
ENSRNOT00000067011
ENSRNOT00000080598 |
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr8_+_71167305 | 5.69 |
ENSRNOT00000021337
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr1_+_72882806 | 5.52 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr3_-_146396299 | 5.39 |
ENSRNOT00000040188
ENSRNOT00000008931 |
Apmap
|
adipocyte plasma membrane associated protein |
| chr6_+_106496992 | 5.38 |
ENSRNOT00000086136
ENSRNOT00000058181 |
Rgs6
|
regulator of G-protein signaling 6 |
| chr12_+_40018937 | 5.32 |
ENSRNOT00000001697
|
Cux2
|
cut-like homeobox 2 |
| chr1_-_170404056 | 5.04 |
ENSRNOT00000024402
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chrX_-_135342996 | 4.97 |
ENSRNOT00000084848
ENSRNOT00000008503 |
Aifm1
|
apoptosis inducing factor, mitochondria associated 1 |
| chr2_-_56679955 | 4.90 |
ENSRNOT00000016722
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr12_+_47698947 | 4.86 |
ENSRNOT00000001586
|
Trpv4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chr5_+_147069616 | 4.77 |
ENSRNOT00000072908
|
Trim62
|
tripartite motif-containing 62 |
| chr9_+_2202511 | 4.62 |
ENSRNOT00000017556
|
Satb1
|
SATB homeobox 1 |
| chr1_-_124803363 | 4.53 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr20_-_14545772 | 4.44 |
ENSRNOT00000001766
|
Bcr
|
BCR, RhoGEF and GTPase activating protein |
| chr8_+_33239139 | 4.20 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr3_-_81304181 | 4.16 |
ENSRNOT00000079746
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr7_+_143810892 | 4.15 |
ENSRNOT00000016240
|
Znf740
|
zinc finger protein 740 |
| chr1_-_265542276 | 4.13 |
ENSRNOT00000065172
|
Mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr6_-_36819821 | 4.08 |
ENSRNOT00000006499
|
Kcns3
|
potassium voltage-gated channel, modifier subfamily S, member 3 |
| chr18_-_3555711 | 3.92 |
ENSRNOT00000029274
|
Tmem241
|
transmembrane protein 241 |
| chr2_+_239415046 | 3.89 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chrX_+_68774699 | 3.78 |
ENSRNOT00000081662
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr1_+_198682230 | 3.62 |
ENSRNOT00000023995
|
Znf48
|
zinc finger protein 48 |
| chr11_+_61531571 | 3.47 |
ENSRNOT00000093467
ENSRNOT00000002727 |
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr5_+_150754021 | 3.44 |
ENSRNOT00000017687
|
Ptafr
|
platelet-activating factor receptor |
| chr10_+_29165577 | 3.31 |
ENSRNOT00000078415
|
Ccnjl
|
cyclin J-like |
| chr7_+_121311024 | 3.29 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr4_+_157836912 | 3.29 |
ENSRNOT00000067271
|
Scnn1a
|
sodium channel epithelial 1 alpha subunit |
| chr14_-_106393670 | 3.24 |
ENSRNOT00000011429
|
Mdh1
|
malate dehydrogenase 1 |
| chr8_+_48443767 | 3.20 |
ENSRNOT00000010127
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr14_+_88549947 | 3.16 |
ENSRNOT00000086177
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr9_+_10471742 | 3.15 |
ENSRNOT00000072276
|
Safb2
|
scaffold attachment factor B2 |
| chr4_-_77706994 | 3.15 |
ENSRNOT00000038517
|
Zfp777
|
zinc finger protein 777 |
| chr7_+_123102493 | 3.15 |
ENSRNOT00000038612
|
Aco2
|
aconitase 2 |
| chrX_+_54734385 | 3.11 |
ENSRNOT00000005023
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr3_+_154905141 | 3.10 |
ENSRNOT00000088748
ENSRNOT00000020556 |
Ralgapb
|
Ral GTPase activating protein non-catalytic beta subunit |
| chr6_-_3355339 | 3.07 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr18_+_30856847 | 3.03 |
ENSRNOT00000027019
|
AABR07031734.1
|
|
| chr7_-_12424367 | 2.97 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr5_+_74766636 | 2.94 |
ENSRNOT00000030913
|
Rn50_5_0791.1
|
|
| chr19_-_42920344 | 2.92 |
ENSRNOT00000019408
ENSRNOT00000068594 |
Zfhx3
|
zinc finger homeobox 3 |
| chr3_-_150156839 | 2.88 |
ENSRNOT00000036217
|
LOC103691939
|
elongation factor 1-alpha 1 pseudogene |
| chr8_+_39878955 | 2.85 |
ENSRNOT00000060300
|
LOC100911068
|
roundabout homolog 4-like |
| chr10_-_90240509 | 2.82 |
ENSRNOT00000028407
|
Atxn7l3
|
ataxin 7-like 3 |
| chr3_+_125320680 | 2.79 |
ENSRNOT00000028891
|
LOC681292
|
hypothetical protein LOC681292 |
| chr3_+_156727642 | 2.72 |
ENSRNOT00000078909
|
Plcg1
|
phospholipase C, gamma 1 |
| chr14_+_81362618 | 2.71 |
ENSRNOT00000017386
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr3_+_123731539 | 2.61 |
ENSRNOT00000051064
|
Cdc25b
|
cell division cycle 25B |
| chr1_-_53087474 | 2.59 |
ENSRNOT00000017302
|
Ccr6
|
C-C motif chemokine receptor 6 |
| chr20_+_3351303 | 2.59 |
ENSRNOT00000080419
ENSRNOT00000001065 ENSRNOT00000086503 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr4_+_145413230 | 2.58 |
ENSRNOT00000056508
|
Il17re
|
interleukin 17 receptor E |
| chr1_-_70003582 | 2.57 |
ENSRNOT00000055721
|
Usp29
|
ubiquitin specific peptidase 29 |
| chr5_-_164348713 | 2.57 |
ENSRNOT00000087044
|
LOC500584
|
similar to casein kinase 1, gamma 3 isoform 2 |
| chr17_-_15094805 | 2.57 |
ENSRNOT00000044073
|
Fbxw17
|
F-box and WD-40 domain protein 17 |
| chr3_+_155297566 | 2.51 |
ENSRNOT00000021435
ENSRNOT00000084866 |
Dhx35
|
DEAH-box helicase 35 |
| chr7_-_70480724 | 2.46 |
ENSRNOT00000090057
|
Dtx3
|
deltex E3 ubiquitin ligase 3 |
| chr1_-_187779675 | 2.44 |
ENSRNOT00000024648
|
Arl6ip1
|
ADP-ribosylation factor like GTPase 6 interacting protein 1 |
| chr10_-_88356538 | 2.43 |
ENSRNOT00000022430
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr6_+_12362813 | 2.40 |
ENSRNOT00000022370
|
Ston1
|
stonin 1 |
| chr11_+_61531416 | 2.35 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr16_-_69242028 | 2.35 |
ENSRNOT00000019002
|
Zfp703
|
zinc finger protein 703 |
| chr10_-_59888198 | 2.35 |
ENSRNOT00000093482
ENSRNOT00000049311 ENSRNOT00000093230 |
Aspa
|
aspartoacylase |
| chr3_-_93789617 | 2.32 |
ENSRNOT00000078616
|
Caprin1
|
cell cycle associated protein 1 |
| chr1_-_221041401 | 2.31 |
ENSRNOT00000064136
|
Pcnx3
|
pecanex homolog 3 (Drosophila) |
| chr10_-_896938 | 2.31 |
ENSRNOT00000086392
|
Nde1
|
nudE neurodevelopment protein 1 |
| chrX_-_74847379 | 2.31 |
ENSRNOT00000067009
|
NEWGENE_1559832
|
ring finger protein, LIM domain interacting |
| chr6_-_76552559 | 2.25 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr16_-_32753278 | 2.24 |
ENSRNOT00000015759
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr20_-_4817146 | 2.23 |
ENSRNOT00000080174
|
Ddx39b
|
DExD-box helicase 39B |
| chr1_-_134870255 | 2.20 |
ENSRNOT00000055829
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr3_-_133894569 | 2.20 |
ENSRNOT00000084221
|
AABR07053968.1
|
|
| chr2_+_28460068 | 2.19 |
ENSRNOT00000066819
|
Foxd1
|
forkhead box D1 |
| chr8_+_36125999 | 2.17 |
ENSRNOT00000013036
ENSRNOT00000088046 |
Kirrel3
|
kin of IRRE like 3 (Drosophila) |
| chr2_-_111793326 | 2.17 |
ENSRNOT00000092660
|
Nlgn1
|
neuroligin 1 |
| chr1_+_221773254 | 2.11 |
ENSRNOT00000028646
|
Rasgrp2
|
RAS guanyl releasing protein 2 |
| chr1_-_73732118 | 2.10 |
ENSRNOT00000077964
|
Leng8
|
leukocyte receptor cluster member 8 |
| chr7_+_120755516 | 2.08 |
ENSRNOT00000056169
|
Kdelr3
|
KDEL endoplasmic reticulum protein retention receptor 3 |
| chr1_+_85460888 | 2.08 |
ENSRNOT00000093384
|
Supt5h
|
SPT5 homolog, DSIF elongation factor subunit |
| chr8_+_39305128 | 2.06 |
ENSRNOT00000008285
|
Fez1
|
fasciculation and elongation protein zeta 1 |
| chr3_+_147609095 | 2.03 |
ENSRNOT00000041456
|
Srxn1
|
sulfiredoxin 1 |
| chr20_-_13657943 | 2.03 |
ENSRNOT00000032290
|
Vpreb3
|
pre-B lymphocyte 3 |
| chr5_-_101138427 | 2.00 |
ENSRNOT00000058615
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr16_-_81243757 | 1.99 |
ENSRNOT00000024677
|
Gas6
|
growth arrest specific 6 |
| chr2_-_113345577 | 1.99 |
ENSRNOT00000034096
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr8_+_48094673 | 1.98 |
ENSRNOT00000008614
|
Nectin1
|
nectin cell adhesion molecule 1 |
| chr3_-_60765645 | 1.97 |
ENSRNOT00000050513
|
Atf2
|
activating transcription factor 2 |
| chr2_-_238803024 | 1.97 |
ENSRNOT00000046775
|
Tet2
|
tet methylcytosine dioxygenase 2 |
| chr10_-_5260608 | 1.92 |
ENSRNOT00000003572
|
Ciita
|
class II, major histocompatibility complex, transactivator |
| chr3_+_113257688 | 1.91 |
ENSRNOT00000019320
|
Map1a
|
microtubule-associated protein 1A |
| chr20_-_5618254 | 1.87 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr10_-_82963919 | 1.86 |
ENSRNOT00000005807
|
Dlx4
|
distal-less homeobox 4 |
| chr8_-_116965396 | 1.86 |
ENSRNOT00000042528
|
Bsn
|
bassoon (presynaptic cytomatrix protein) |
| chr13_-_78521911 | 1.85 |
ENSRNOT00000087506
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr9_-_93404883 | 1.78 |
ENSRNOT00000025024
|
Nmur1
|
neuromedin U receptor 1 |
| chr1_+_80954858 | 1.76 |
ENSRNOT00000030440
|
Zfp112
|
zinc finger protein 112 |
| chr1_-_91588609 | 1.72 |
ENSRNOT00000050931
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr10_-_88357025 | 1.69 |
ENSRNOT00000080006
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr3_+_14304591 | 1.68 |
ENSRNOT00000007776
|
Cntrl
|
centriolin |
| chr15_-_37030240 | 1.62 |
ENSRNOT00000028219
ENSRNOT00000088037 |
Pspc1
|
paraspeckle component 1 |
| chr15_-_86142672 | 1.61 |
ENSRNOT00000057830
|
Commd6
|
COMM domain containing 6 |
| chr20_+_29831314 | 1.54 |
ENSRNOT00000000696
|
Psap
|
prosaposin |
| chr2_+_38230757 | 1.50 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chr1_-_213973163 | 1.50 |
ENSRNOT00000024867
|
LOC100911402
|
cell cycle exit and neuronal differentiation protein 1-like |
| chr7_+_73273985 | 1.43 |
ENSRNOT00000077730
|
Pop1
|
POP1 homolog, ribonuclease P/MRP subunit |
| chr14_+_76732650 | 1.42 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr5_+_138300107 | 1.40 |
ENSRNOT00000047151
|
Cldn19
|
claudin 19 |
| chr4_+_152449270 | 1.39 |
ENSRNOT00000076733
|
Rad52
|
RAD52 homolog, DNA repair protein |
| chr14_-_44375804 | 1.36 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr10_+_61685241 | 1.35 |
ENSRNOT00000092606
|
Mnt
|
MAX network transcriptional repressor |
| chr8_+_114867062 | 1.30 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr8_+_29453643 | 1.30 |
ENSRNOT00000090643
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr17_+_86066833 | 1.24 |
ENSRNOT00000022661
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr1_+_87066289 | 1.23 |
ENSRNOT00000027645
|
Capn12
|
calpain 12 |
| chr1_-_78968329 | 1.22 |
ENSRNOT00000023078
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr5_+_144701949 | 1.20 |
ENSRNOT00000015618
|
Psmb2
|
proteasome subunit beta 2 |
| chr1_+_61374379 | 1.19 |
ENSRNOT00000015343
|
Zfp53
|
zinc finger protein 53 |
| chr8_-_133128290 | 1.19 |
ENSRNOT00000008783
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr12_-_30566032 | 1.17 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr10_+_59000397 | 1.14 |
ENSRNOT00000021134
|
Mybbp1a
|
MYB binding protein 1a |
| chr20_+_28572242 | 1.09 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr18_-_31430973 | 1.08 |
ENSRNOT00000026065
|
Pcdh12
|
protocadherin 12 |
| chr1_+_224882439 | 1.00 |
ENSRNOT00000024785
|
Chrm1
|
cholinergic receptor, muscarinic 1 |
| chr1_-_64175099 | 1.00 |
ENSRNOT00000091288
|
Ndufa3
|
NADH:ubiquinone oxidoreductase subunit A3 |
| chr10_-_104748003 | 0.98 |
ENSRNOT00000042372
ENSRNOT00000046754 |
Acox1
|
acyl-CoA oxidase 1 |
| chr4_-_145487426 | 0.94 |
ENSRNOT00000013488
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr3_-_153952108 | 0.94 |
ENSRNOT00000050465
|
Adig
|
adipogenin |
| chr10_-_15155412 | 0.91 |
ENSRNOT00000026536
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr10_+_82032656 | 0.87 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr17_-_90315492 | 0.86 |
ENSRNOT00000070807
|
Gng4
|
G protein subunit gamma 4 |
| chr18_+_70733872 | 0.85 |
ENSRNOT00000067018
|
Acaa2
|
acetyl-CoA acyltransferase 2 |
| chr6_-_143195143 | 0.85 |
ENSRNOT00000081337
|
AABR07065837.1
|
|
| chr17_+_45247776 | 0.83 |
ENSRNOT00000081063
ENSRNOT00000080655 |
Zkscan3
|
zinc finger with KRAB and SCAN domains 3 |
| chr10_-_14247886 | 0.75 |
ENSRNOT00000020504
|
Nubp2
|
nucleotide binding protein 2 |
| chr13_-_71282802 | 0.75 |
ENSRNOT00000025544
|
Rgsl2h
|
regulator of G-protein signaling like 2 homolog (mouse) |
| chr10_+_110893232 | 0.74 |
ENSRNOT00000072190
|
Metrnl
|
meteorin-like, glial cell differentiation regulator |
| chr6_-_135049728 | 0.74 |
ENSRNOT00000009556
|
Hsp90aa1
|
heat shock protein 90 alpha family class A member 1 |
| chr12_-_46718355 | 0.72 |
ENSRNOT00000030031
|
Rab35
|
RAB35, member RAS oncogene family |
| chr20_+_9586075 | 0.70 |
ENSRNOT00000001527
ENSRNOT00000079509 |
Glp1r
|
glucagon-like peptide 1 receptor |
| chr8_+_104106740 | 0.66 |
ENSRNOT00000015015
|
Tfdp2
|
transcription factor Dp-2 |
| chr6_-_107678156 | 0.62 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr10_-_85517683 | 0.61 |
ENSRNOT00000016070
|
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr3_+_117853999 | 0.59 |
ENSRNOT00000042949
|
LOC100911847
|
40S ribosomal protein S14-like |
| chr1_-_219853329 | 0.58 |
ENSRNOT00000026169
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr4_+_82637411 | 0.57 |
ENSRNOT00000048225
|
LOC685406
|
LRRGT00062 |
| chr13_-_85622314 | 0.57 |
ENSRNOT00000005719
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr7_+_98432024 | 0.53 |
ENSRNOT00000091077
|
Fer1l6
|
fer-1-like family member 6 |
| chr12_+_30441055 | 0.53 |
ENSRNOT00000081099
|
AABR07036007.1
|
|
| chr4_+_170958196 | 0.52 |
ENSRNOT00000007905
|
Pde6h
|
phosphodiesterase 6H |
| chrX_-_78911601 | 0.52 |
ENSRNOT00000003188
|
RGD1566265
|
similar to RIKEN cDNA 2610002M06 |
| chr1_-_62191818 | 0.50 |
ENSRNOT00000033187
|
AABR07001926.1
|
|
| chr3_+_73366155 | 0.49 |
ENSRNOT00000044979
|
Olr473
|
olfactory receptor 473 |
| chr8_-_48449311 | 0.42 |
ENSRNOT00000010134
|
Rnf26
|
ring finger protein 26 |
| chr8_+_59164572 | 0.41 |
ENSRNOT00000015102
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr1_-_170927957 | 0.38 |
ENSRNOT00000049686
|
Olr218
|
olfactory receptor 218 |
| chr1_-_126211439 | 0.36 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr6_-_143195445 | 0.35 |
ENSRNOT00000078672
|
AABR07065837.1
|
|
| chr18_-_63185510 | 0.34 |
ENSRNOT00000024632
|
Afg3l2
|
AFG3 like matrix AAA peptidase subunit 2 |
| chr14_-_105069565 | 0.34 |
ENSRNOT00000067365
|
AABR07016578.1
|
|
| chr10_+_29123273 | 0.30 |
ENSRNOT00000005171
|
Ccnjl
|
cyclin J-like |
| chr3_+_94549180 | 0.27 |
ENSRNOT00000016318
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr5_-_59149625 | 0.23 |
ENSRNOT00000040641
ENSRNOT00000084216 |
Spag8
|
sperm associated antigen 8 |
| chr7_-_114590119 | 0.21 |
ENSRNOT00000079599
|
Ptk2
|
protein tyrosine kinase 2 |
| chr2_+_252305874 | 0.17 |
ENSRNOT00000020972
|
Ctbs
|
chitobiase |
| chr18_+_29587901 | 0.12 |
ENSRNOT00000085470
|
Wdr55
|
WD repeat domain 55 |
| chr19_+_25095089 | 0.11 |
ENSRNOT00000041717
|
Prkaca
|
protein kinase cAMP-activated catalytic subunit alpha |
| chrX_-_113474164 | 0.08 |
ENSRNOT00000046804
|
Gucy2f
|
guanylate cyclase 2F |
| chr3_-_60813869 | 0.08 |
ENSRNOT00000058234
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr16_+_1979191 | 0.06 |
ENSRNOT00000014382
|
Ppif
|
peptidylprolyl isomerase F |
| chr8_-_40122450 | 0.04 |
ENSRNOT00000039729
|
Tbrg1
|
transforming growth factor beta regulator 1 |
| chr3_+_21313227 | 0.01 |
ENSRNOT00000036335
|
LOC690140
|
similar to olfactory receptor Olr1374 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rora
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.8 | GO:0007493 | endodermal cell fate determination(GO:0007493) negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 3.3 | 10.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 3.1 | 9.2 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 2.1 | 6.4 | GO:0015820 | leucine transport(GO:0015820) |
| 2.1 | 12.5 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 2.0 | 6.1 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) protein import into mitochondrial intermembrane space(GO:0045041) |
| 2.0 | 6.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 1.6 | 4.9 | GO:0072566 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 1.5 | 4.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 1.3 | 10.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.2 | 6.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 1.1 | 3.4 | GO:0043313 | response to symbiotic bacterium(GO:0009609) regulation of neutrophil degranulation(GO:0043313) |
| 1.0 | 4.1 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 1.0 | 9.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 1.0 | 5.0 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.9 | 2.6 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.8 | 2.4 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.8 | 3.9 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.8 | 3.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.8 | 2.3 | GO:1900095 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.7 | 2.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.7 | 7.7 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.7 | 2.7 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.7 | 2.0 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.6 | 2.6 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.6 | 4.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.6 | 7.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.6 | 3.0 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.6 | 3.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.6 | 7.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.5 | 2.2 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) pattern specification involved in metanephros development(GO:0072268) |
| 0.5 | 2.2 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.5 | 2.2 | GO:0099545 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.5 | 3.2 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.5 | 2.0 | GO:0097018 | cellular response to vitamin K(GO:0071307) renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.5 | 2.3 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.5 | 2.7 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.4 | 5.7 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.4 | 7.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.4 | 7.0 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.4 | 5.3 | GO:0007614 | short-term memory(GO:0007614) |
| 0.4 | 4.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 7.9 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.4 | 2.6 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.4 | 1.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.3 | 5.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 2.0 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.3 | 4.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.3 | 1.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.3 | 4.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 | 1.9 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.3 | 2.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.3 | 1.9 | GO:0002339 | B cell selection(GO:0002339) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.3 | 1.9 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.3 | 2.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 2.0 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.3 | 3.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.3 | 1.5 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.2 | 0.7 | GO:0061741 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 0.2 | 1.4 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.2 | 2.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 2.0 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.2 | 2.4 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.2 | 3.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 8.5 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.2 | 1.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 4.8 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.2 | 0.9 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 6.9 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.2 | 3.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 5.9 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.2 | 2.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.2 | 0.9 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.2 | 1.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 1.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 2.8 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 6.6 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.1 | 0.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 2.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 3.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.4 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 5.0 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 1.4 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 2.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 3.2 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 9.7 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.1 | 0.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 0.4 | GO:0071000 | response to magnetism(GO:0071000) |
| 0.1 | 19.9 | GO:0001889 | liver development(GO:0001889) |
| 0.1 | 0.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 2.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 3.3 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.1 | 1.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.8 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 1.2 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 1.8 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 1.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 4.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 6.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.6 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 1.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 3.9 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 2.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 2.3 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 0.5 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 9.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 1.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.3 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 1.3 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 2.4 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 1.5 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.0 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 14.6 | GO:0008091 | spectrin(GO:0008091) cuticular plate(GO:0032437) |
| 1.4 | 4.2 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 1.4 | 5.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 1.3 | 5.0 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 1.2 | 6.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.1 | 6.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.1 | 3.2 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.6 | 2.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.6 | 1.9 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.5 | 2.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.5 | 2.4 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.5 | 5.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.5 | 3.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.5 | 1.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.4 | 6.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 4.0 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.3 | 3.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 1.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.2 | 2.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 4.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 3.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 2.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 6.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 6.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.4 | GO:0045242 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 0.1 | 3.3 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 2.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 1.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 9.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 10.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 21.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 4.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 2.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 11.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 6.5 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 3.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 4.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 2.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 2.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 9.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 18.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 8.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 2.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 8.1 | GO:0098794 | postsynapse(GO:0098794) |
| 0.0 | 2.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 4.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 18.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.7 | 3.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 1.4 | 8.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.4 | 5.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 1.2 | 6.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.0 | 3.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.9 | 2.7 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.8 | 4.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.8 | 3.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.8 | 2.4 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.8 | 3.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.8 | 5.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.7 | 6.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.7 | 10.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.7 | 3.9 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.6 | 11.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.6 | 5.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.5 | 4.9 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.5 | 3.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.5 | 3.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.5 | 2.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.5 | 1.4 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.4 | 2.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 3.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.4 | 12.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.4 | 2.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.4 | 2.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.4 | 3.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 12.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 2.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 2.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.3 | 2.2 | GO:0017070 | U6 snRNA binding(GO:0017070) U4 snRNA binding(GO:0030621) |
| 0.3 | 5.8 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 4.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 6.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.2 | 6.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 6.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 1.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 1.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 1.0 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 1.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 1.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 4.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 0.9 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 0.7 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 11.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 2.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 14.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 6.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.9 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.1 | 1.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 33.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 2.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 21.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 6.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 14.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 5.1 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 2.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 1.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 1.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.9 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 2.5 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.5 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 2.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 2.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 2.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 6.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 6.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 2.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 3.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 6.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 12.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 10.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 5.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 4.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 10.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 6.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 7.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 7.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 3.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 11.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 2.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 5.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 29.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.6 | 2.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.6 | 10.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.6 | 16.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.4 | 8.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.4 | 6.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 11.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 5.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 3.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 2.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 1.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 5.6 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 2.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 12.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 4.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 6.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 6.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 3.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 10.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 5.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 3.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.6 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 2.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |