Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
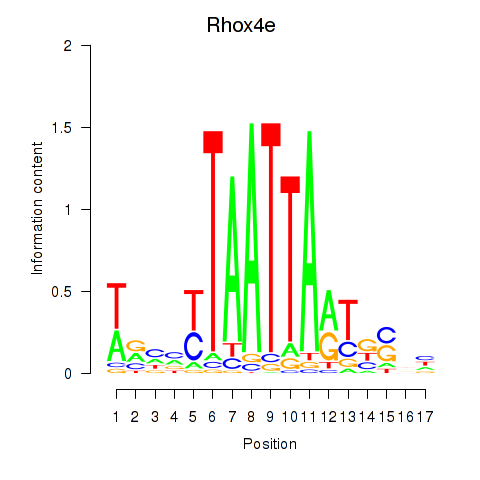
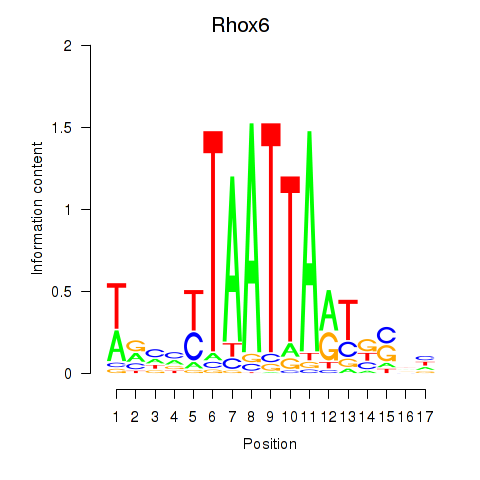
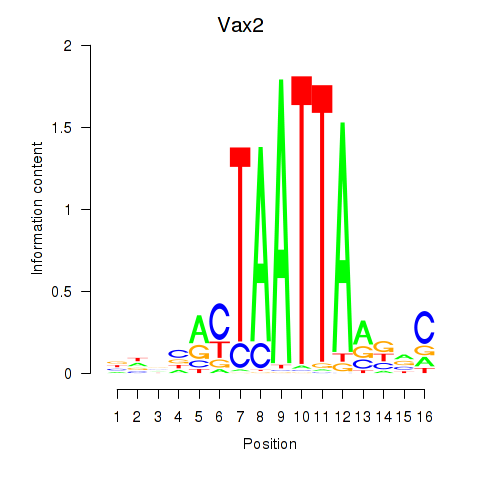
Results for Rhox4e_Rhox6_Vax2
Z-value: 0.78
Transcription factors associated with Rhox4e_Rhox6_Vax2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Vax2
|
ENSRNOG00000013261 | ventral anterior homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Psx1 | rn6_v1_chr3_-_574592_574592 | -0.04 | 4.5e-01 | Click! |
| Vax2 | rn6_v1_chr4_+_115388839_115388839 | -0.02 | 7.5e-01 | Click! |
Activity profile of Rhox4e_Rhox6_Vax2 motif
Sorted Z-values of Rhox4e_Rhox6_Vax2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_14229067 | 70.09 |
ENSRNOT00000025534
ENSRNOT00000092865 |
C5
|
complement C5 |
| chr3_+_159368273 | 55.63 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr7_-_71139267 | 51.85 |
ENSRNOT00000065232
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr3_+_171213936 | 48.11 |
ENSRNOT00000031586
|
Pck1
|
phosphoenolpyruvate carboxykinase 1 |
| chr3_+_159902441 | 34.63 |
ENSRNOT00000089893
ENSRNOT00000011978 |
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr3_+_95233874 | 33.82 |
ENSRNOT00000079743
|
LOC691083
|
hypothetical protein LOC691083 |
| chr6_+_56846789 | 28.89 |
ENSRNOT00000032108
|
Agmo
|
alkylglycerol monooxygenase |
| chr1_+_48273611 | 22.88 |
ENSRNOT00000022254
ENSRNOT00000022068 |
Slc22a1
|
solute carrier family 22 member 1 |
| chr18_-_28425944 | 22.70 |
ENSRNOT00000084372
|
Slc23a1
|
solute carrier family 23 member 1 |
| chr14_+_22517774 | 21.91 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr2_+_158097843 | 21.81 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr14_+_22806132 | 21.61 |
ENSRNOT00000002728
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr16_+_74865516 | 21.13 |
ENSRNOT00000058072
|
Atp7b
|
ATPase copper transporting beta |
| chrX_-_23144324 | 19.95 |
ENSRNOT00000000178
ENSRNOT00000081239 |
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr15_-_42693694 | 18.50 |
ENSRNOT00000022702
|
Gulo
|
gulonolactone (L-) oxidase |
| chr13_-_83457888 | 18.29 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr16_+_10267482 | 17.90 |
ENSRNOT00000085255
|
Gdf2
|
growth differentiation factor 2 |
| chr6_-_7058314 | 17.51 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr6_+_2216623 | 17.07 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chrX_-_110232179 | 16.34 |
ENSRNOT00000014739
|
Serpina7
|
serpin family A member 7 |
| chr10_+_43601689 | 15.62 |
ENSRNOT00000029238
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr8_+_106449321 | 15.32 |
ENSRNOT00000018622
|
Rbp1
|
retinol binding protein 1 |
| chr9_-_4978892 | 14.46 |
ENSRNOT00000015189
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr2_-_181900856 | 14.32 |
ENSRNOT00000082156
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr1_-_224698514 | 14.05 |
ENSRNOT00000024234
|
Slc22a25
|
solute carrier family 22, member 25 |
| chr17_+_32082937 | 13.71 |
ENSRNOT00000074775
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr17_+_9736786 | 13.45 |
ENSRNOT00000081920
|
F12
|
coagulation factor XII |
| chr1_+_185863043 | 13.27 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr7_-_69982592 | 13.06 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr20_+_4593389 | 12.85 |
ENSRNOT00000001174
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr1_+_88113445 | 12.25 |
ENSRNOT00000056955
|
Ggn
|
gametogenetin |
| chr2_+_54466280 | 11.73 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr16_+_29674793 | 11.59 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr17_+_69468427 | 11.37 |
ENSRNOT00000058413
|
RGD1564865
|
similar to 20-alpha-hydroxysteroid dehydrogenase |
| chr1_+_229100752 | 11.04 |
ENSRNOT00000074881
|
LOC686967
|
similar to olfactory receptor 1442 |
| chr7_-_143967484 | 10.84 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr4_-_100252755 | 10.60 |
ENSRNOT00000017301
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr9_-_79630452 | 10.31 |
ENSRNOT00000078125
ENSRNOT00000086044 ENSRNOT00000089283 |
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr11_+_74057361 | 10.21 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chr19_-_50220455 | 10.21 |
ENSRNOT00000079760
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr18_+_81694808 | 9.92 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chr1_-_276228574 | 9.91 |
ENSRNOT00000021746
|
Gucy2g
|
guanylate cyclase 2G |
| chr1_-_198104109 | 9.84 |
ENSRNOT00000026186
|
Sult1a1
|
sulfotransferase family 1A member 1 |
| chr8_+_70994563 | 9.75 |
ENSRNOT00000051504
ENSRNOT00000077163 |
Spg21
|
spastic paraplegia 21 homolog (human) |
| chr16_-_75241303 | 9.54 |
ENSRNOT00000058056
|
Defb2
|
defensin beta 2 |
| chr8_+_85059051 | 9.32 |
ENSRNOT00000033196
|
Gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr7_-_126382449 | 9.25 |
ENSRNOT00000085540
|
7530416G11Rik
|
RIKEN cDNA 7530416G11 gene |
| chr14_-_60276794 | 9.21 |
ENSRNOT00000048509
|
Slc34a2
|
solute carrier family 34 member 2 |
| chr13_-_50514151 | 9.11 |
ENSRNOT00000003951
|
Ren
|
renin |
| chr1_-_167971151 | 9.09 |
ENSRNOT00000043023
|
Olr53
|
olfactory receptor 53 |
| chr4_+_82665094 | 9.09 |
ENSRNOT00000078254
|
AABR07060591.1
|
|
| chr5_+_154294806 | 9.07 |
ENSRNOT00000012853
|
Hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chrX_-_32095867 | 8.60 |
ENSRNOT00000049947
ENSRNOT00000080730 |
Ace2
|
angiotensin I converting enzyme 2 |
| chr14_-_84334066 | 8.52 |
ENSRNOT00000006160
|
Mtfp1
|
mitochondrial fission process 1 |
| chr13_+_90943255 | 8.37 |
ENSRNOT00000011539
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr10_-_106883423 | 8.18 |
ENSRNOT00000071406
|
LOC688286
|
similar to threonine aldolase 1 |
| chr5_-_147412705 | 8.07 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr4_+_88694583 | 7.91 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chrM_+_5323 | 7.88 |
ENSRNOT00000050156
|
Mt-co1
|
mitochondrially encoded cytochrome c oxidase 1 |
| chr5_-_119564846 | 7.80 |
ENSRNOT00000012977
|
Cyp2j4
|
cytochrome P450, family 2, subfamily j, polypeptide 4 |
| chr2_+_80269661 | 7.71 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chr14_-_2032593 | 7.71 |
ENSRNOT00000000037
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr8_-_7426611 | 7.51 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr5_+_165465095 | 7.47 |
ENSRNOT00000031978
|
LOC691354
|
hypothetical protein LOC691354 |
| chr4_+_148782479 | 7.38 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr1_-_22625204 | 7.11 |
ENSRNOT00000021694
|
Vnn1
|
vanin 1 |
| chr3_-_165537940 | 7.07 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr9_+_20048121 | 7.03 |
ENSRNOT00000014791
|
Mep1a
|
meprin 1 subunit alpha |
| chr3_-_76135475 | 7.01 |
ENSRNOT00000007712
|
Olr604
|
olfactory receptor 604 |
| chr4_+_52235811 | 6.91 |
ENSRNOT00000060478
|
Hyal5
|
hyaluronoglucosaminidase 5 |
| chr16_-_24951612 | 6.34 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chrM_+_11736 | 6.19 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr18_+_4084228 | 6.05 |
ENSRNOT00000071392
|
Cabyr
|
calcium binding tyrosine phosphorylation regulated |
| chr8_-_113689681 | 6.04 |
ENSRNOT00000056435
|
LOC688828
|
similar to Nucleoside diphosphate-linked moiety X motif 16 (Nudix motif 16) |
| chr14_+_23405717 | 5.99 |
ENSRNOT00000029805
|
Tmprss11c
|
transmembrane protease, serine 11C |
| chr17_+_38323674 | 5.97 |
ENSRNOT00000021982
|
Prl5a1
|
prolactin family 5, subfamily a, member 1 |
| chrM_+_7006 | 5.84 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr10_-_87564327 | 5.79 |
ENSRNOT00000064760
ENSRNOT00000068237 |
LOC680160
|
similar to keratin associated protein 4-7 |
| chr20_-_13994794 | 5.74 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr10_-_47546345 | 5.55 |
ENSRNOT00000077461
|
Aldh3a2
|
aldehyde dehydrogenase 3 family, member A2 |
| chr10_-_47546037 | 5.45 |
ENSRNOT00000066109
|
Aldh3a2
|
aldehyde dehydrogenase 3 family, member A2 |
| chr10_-_88670430 | 5.29 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr3_-_74998096 | 5.27 |
ENSRNOT00000051768
|
LOC100910165
|
olfactory receptor 1052-like |
| chr1_+_13595295 | 5.24 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr8_+_117117430 | 5.16 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr12_+_24761210 | 5.07 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr10_+_103206014 | 5.05 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr2_-_158156444 | 4.94 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr14_+_18983853 | 4.90 |
ENSRNOT00000003836
|
Rassf6
|
Ras association domain family member 6 |
| chr6_+_104291071 | 4.66 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr8_-_43304560 | 4.63 |
ENSRNOT00000060092
|
Olr1307
|
olfactory receptor 1307 |
| chr3_+_73235594 | 4.51 |
ENSRNOT00000082357
|
Olr463
|
olfactory receptor 463 |
| chrX_+_74200972 | 4.41 |
ENSRNOT00000076956
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chrX_-_124252447 | 4.37 |
ENSRNOT00000061546
|
Rhox12
|
reproductive homeobox 12 |
| chr20_-_9855443 | 4.30 |
ENSRNOT00000090275
ENSRNOT00000066266 |
Tff3
|
trefoil factor 3 |
| chrM_+_2740 | 4.27 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr1_-_15374850 | 4.26 |
ENSRNOT00000016728
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chr2_-_132301073 | 4.17 |
ENSRNOT00000058288
|
RGD1563562
|
similar to GTPase activating protein testicular GAP1 |
| chr6_-_102372611 | 4.14 |
ENSRNOT00000085927
|
Rdh11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr4_+_1470716 | 4.13 |
ENSRNOT00000044223
|
Olr1235
|
olfactory receptor 1235 |
| chr6_+_104291340 | 4.09 |
ENSRNOT00000089313
|
Slc39a9
|
solute carrier family 39, member 9 |
| chrX_+_74205842 | 4.07 |
ENSRNOT00000077003
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr10_+_56445647 | 4.05 |
ENSRNOT00000056870
|
Tmem256
|
transmembrane protein 256 |
| chr5_+_25253010 | 4.04 |
ENSRNOT00000061333
|
Rbm12b
|
RNA binding motif protein 12B |
| chr2_+_188583664 | 4.02 |
ENSRNOT00000045477
|
Dpm3
|
dolichyl-phosphate mannosyltransferase subunit 3 |
| chr18_+_45023932 | 4.02 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chrX_-_111102464 | 3.96 |
ENSRNOT00000084176
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr5_+_25725683 | 3.89 |
ENSRNOT00000087602
|
LOC679087
|
similar to swan |
| chr2_+_127525285 | 3.84 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chrX_+_37329779 | 3.80 |
ENSRNOT00000038352
ENSRNOT00000088802 |
Pdha1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr3_+_1413671 | 3.78 |
ENSRNOT00000007675
|
Il1f10
|
interleukin 1 family member 10 |
| chr1_+_128637049 | 3.64 |
ENSRNOT00000018639
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr14_+_6221085 | 3.64 |
ENSRNOT00000068215
|
AABR07014259.1
|
|
| chr9_-_121972055 | 3.53 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr1_+_220470089 | 3.52 |
ENSRNOT00000027406
ENSRNOT00000078657 |
Yif1a
|
Yip1 interacting factor homolog A, membrane trafficking protein |
| chr2_-_61692487 | 3.51 |
ENSRNOT00000078544
|
LOC499541
|
LRRGT00045 |
| chr16_+_51748970 | 3.50 |
ENSRNOT00000059182
|
Adam26a
|
a disintegrin and metallopeptidase domain 26A |
| chr14_-_6900733 | 3.47 |
ENSRNOT00000061224
|
Dmp1
|
dentin matrix acidic phosphoprotein 1 |
| chrX_+_136715598 | 3.47 |
ENSRNOT00000040628
|
Olfr1320
|
olfactory receptor 1320 |
| chr10_-_60032218 | 3.40 |
ENSRNOT00000072328
|
Olr1481
|
olfactory receptor 1481 |
| chr10_-_82117109 | 3.39 |
ENSRNOT00000079711
|
Abcc3
|
ATP binding cassette subfamily C member 3 |
| chr8_-_43443531 | 3.28 |
ENSRNOT00000007901
|
LOC103693060
|
olfactory receptor 148-like |
| chr14_-_16903242 | 3.26 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr14_+_34446616 | 3.26 |
ENSRNOT00000002976
|
Clock
|
clock circadian regulator |
| chr3_-_75576520 | 3.22 |
ENSRNOT00000083330
|
Olr567
|
olfactory receptor 567 |
| chr18_+_79773608 | 3.22 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr8_+_42260375 | 3.21 |
ENSRNOT00000074713
|
LOC100912245
|
olfactory receptor 143-like |
| chr10_-_87286387 | 3.20 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr1_-_14117021 | 3.19 |
ENSRNOT00000004344
|
RGD1560303
|
similar to hypothetical protein 4933423E17 |
| chr17_-_79829511 | 3.18 |
ENSRNOT00000085909
|
AABR07028668.1
|
|
| chr10_-_78219793 | 3.15 |
ENSRNOT00000003369
|
Tom1l1
|
target of myb1 like 1 membrane trafficking protein |
| chr2_+_127489771 | 3.04 |
ENSRNOT00000093581
|
Intu
|
inturned planar cell polarity protein |
| chr14_-_70160624 | 3.01 |
ENSRNOT00000022589
|
Clrn2
|
clarin 2 |
| chr1_-_149675443 | 2.96 |
ENSRNOT00000086084
|
Olr294
|
olfactory receptor 294 |
| chr3_-_74991168 | 2.93 |
ENSRNOT00000049273
|
Olr550
|
olfactory receptor 550 |
| chr1_+_214183724 | 2.92 |
ENSRNOT00000091150
|
Lrrc56
|
leucine rich repeat containing 56 |
| chr4_+_147832136 | 2.90 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr7_+_63747577 | 2.89 |
ENSRNOT00000086123
ENSRNOT00000067395 ENSRNOT00000080052 |
RGD1561648
|
RGD1561648 |
| chr6_-_104290579 | 2.86 |
ENSRNOT00000066014
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr16_+_7035068 | 2.85 |
ENSRNOT00000024296
|
Nek4
|
NIMA-related kinase 4 |
| chrX_-_105308554 | 2.84 |
ENSRNOT00000045556
|
Taf7l
|
TATA-box binding protein associated factor 7-like |
| chr13_+_84474319 | 2.82 |
ENSRNOT00000031367
ENSRNOT00000072244 ENSRNOT00000072897 ENSRNOT00000064168 ENSRNOT00000074954 ENSRNOT00000073696 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr3_+_70327193 | 2.80 |
ENSRNOT00000089165
|
Fsip2
|
fibrous sheath-interacting protein 2 |
| chr19_+_52258947 | 2.79 |
ENSRNOT00000021072
|
Adad2
|
adenosine deaminase domain containing 2 |
| chr10_-_41491554 | 2.74 |
ENSRNOT00000035759
|
LOC102552619
|
uncharacterized LOC102552619 |
| chr2_-_216382244 | 2.74 |
ENSRNOT00000086695
ENSRNOT00000087259 |
LOC103689940
|
pancreatic alpha-amylase-like |
| chr3_+_73249710 | 2.72 |
ENSRNOT00000083865
|
Olr464
|
olfactory receptor 464 |
| chr2_+_252090669 | 2.68 |
ENSRNOT00000020656
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr18_-_43945273 | 2.66 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr3_+_76237607 | 2.61 |
ENSRNOT00000067444
|
Olr606
|
olfactory receptor 606 |
| chr20_-_34679779 | 2.60 |
ENSRNOT00000078420
|
Cep85l
|
centrosomal protein 85-like |
| chr17_-_30661690 | 2.59 |
ENSRNOT00000048146
|
RGD1307537
|
similar to RIKEN cDNA 4933417A18 |
| chr1_-_134871167 | 2.59 |
ENSRNOT00000076300
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr10_-_34301197 | 2.57 |
ENSRNOT00000044667
|
Olr1383
|
olfactory receptor 1383 |
| chr16_+_72139063 | 2.56 |
ENSRNOT00000023805
|
Adam18
|
ADAM metallopeptidase domain 18 |
| chr1_+_80141630 | 2.56 |
ENSRNOT00000029552
|
Opa3
|
optic atrophy 3 |
| chr2_+_240642847 | 2.56 |
ENSRNOT00000079694
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_88684415 | 2.53 |
ENSRNOT00000009001
|
LOC500148
|
similar to 40S ribosomal protein S7 (S8) |
| chr14_-_6533524 | 2.52 |
ENSRNOT00000079795
|
Abcg3l1
|
ATP-binding cassette, subfamily G (WHITE), member 3-like 1 |
| chr7_+_99609191 | 2.51 |
ENSRNOT00000012883
|
Sqle
|
squalene epoxidase |
| chr1_+_201337416 | 2.47 |
ENSRNOT00000067742
|
Btbd16
|
BTB domain containing 16 |
| chr7_+_144531814 | 2.40 |
ENSRNOT00000033300
|
Hoxc13
|
homeobox C13 |
| chr3_-_165700489 | 2.35 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr3_+_15560712 | 2.35 |
ENSRNOT00000010218
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr1_-_148847662 | 2.34 |
ENSRNOT00000090329
|
LOC100911376
|
olfactory receptor 226-like |
| chr5_+_28485619 | 2.29 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr1_-_173456488 | 2.26 |
ENSRNOT00000044753
|
Olr282
|
olfactory receptor 282 |
| chr15_+_108318664 | 2.23 |
ENSRNOT00000057469
|
Ubac2
|
UBA domain containing 2 |
| chr9_-_119190698 | 2.20 |
ENSRNOT00000021534
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr7_-_5168902 | 2.18 |
ENSRNOT00000090661
|
Olr894
|
olfactory receptor 894 |
| chr8_-_105226588 | 2.18 |
ENSRNOT00000071936
|
Trim42
|
tripartite motif-containing 42 |
| chr14_-_44078897 | 2.16 |
ENSRNOT00000031792
|
N4bp2
|
NEDD4 binding protein 2 |
| chr13_-_84795070 | 2.15 |
ENSRNOT00000076083
ENSRNOT00000029706 |
Fmo9
|
flavin containing monooxygenase 9 |
| chr4_-_165747201 | 2.13 |
ENSRNOT00000007435
|
Tas2r130
|
taste receptor, type 2, member 130 |
| chr10_-_87521514 | 2.11 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr1_-_149603272 | 2.08 |
ENSRNOT00000045582
|
Olr13
|
olfactory receptor 13 |
| chrX_+_45614888 | 2.06 |
ENSRNOT00000058473
|
LOC100909677
|
uncharacterized LOC100909677 |
| chr3_-_30930141 | 2.05 |
ENSRNOT00000037001
|
AABR07051982.1
|
|
| chr7_+_78732904 | 2.03 |
ENSRNOT00000088433
|
LOC103690326
|
dendritic cell-specific transmembrane protein |
| chr1_-_227457629 | 2.03 |
ENSRNOT00000035910
ENSRNOT00000073770 |
Ms4a5
|
membrane spanning 4-domains A5 |
| chr1_+_253221812 | 1.99 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chr7_+_32193086 | 1.95 |
ENSRNOT00000079193
|
AABR07056611.1
|
|
| chr3_-_77494108 | 1.94 |
ENSRNOT00000047726
|
Olr662
|
olfactory receptor 662 |
| chr8_+_18785118 | 1.94 |
ENSRNOT00000090997
|
LOC686748
|
similar to olfactory receptor 829 |
| chr1_-_170694872 | 1.92 |
ENSRNOT00000075443
|
LOC100911839
|
olfactory receptor 2D3-like |
| chr4_+_162937774 | 1.90 |
ENSRNOT00000083188
|
Clec2g
|
C-type lectin domain family 2, member G |
| chr5_-_110978644 | 1.90 |
ENSRNOT00000090548
|
AABR07049228.1
|
|
| chr3_+_55461420 | 1.89 |
ENSRNOT00000073549
|
G6pc2
|
glucose-6-phosphatase catalytic subunit 2 |
| chr4_-_177039677 | 1.86 |
ENSRNOT00000055538
ENSRNOT00000075667 |
RGD1561551
|
similar to Hypothetical protein MGC75664 |
| chr2_-_60657712 | 1.84 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr3_-_75089064 | 1.80 |
ENSRNOT00000046167
|
Olr542
|
olfactory receptor 542 |
| chr3_+_75957592 | 1.80 |
ENSRNOT00000073455
|
Olr590
|
olfactory receptor 590 |
| chr1_-_230687157 | 1.75 |
ENSRNOT00000048193
|
Olr380
|
olfactory receptor 380 |
| chrX_+_107584653 | 1.75 |
ENSRNOT00000072037
|
AABR07040686.1
|
|
| chr3_-_75720299 | 1.73 |
ENSRNOT00000071529
|
Olr576
|
olfactory receptor 576 |
| chr2_+_148765412 | 1.71 |
ENSRNOT00000018248
ENSRNOT00000084662 |
Selenot
|
selenoprotein T |
| chr3_-_75235439 | 1.70 |
ENSRNOT00000046688
|
Olr552
|
olfactory receptor 552 |
| chr4_-_156563438 | 1.70 |
ENSRNOT00000045305
|
Vom2r51
|
vomeronasal 2 receptor, 51 |
| chr2_-_216348194 | 1.69 |
ENSRNOT00000087839
|
LOC103689940
|
pancreatic alpha-amylase-like |
| chr13_-_78885464 | 1.60 |
ENSRNOT00000003828
|
Dars2
|
aspartyl-tRNA synthetase 2 (mitochondrial) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rhox4e_Rhox6_Vax2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.0 | 48.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 11.7 | 70.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 8.7 | 34.6 | GO:0042977 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 7.6 | 22.7 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 7.1 | 21.3 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 7.0 | 21.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 5.5 | 21.8 | GO:1903015 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 4.6 | 18.5 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 4.5 | 13.5 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 4.4 | 17.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 3.9 | 11.7 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 3.7 | 29.6 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 3.5 | 17.7 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 3.5 | 10.6 | GO:1903595 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) positive regulation of histamine secretion by mast cell(GO:1903595) |
| 2.8 | 33.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 2.7 | 35.7 | GO:1901374 | acetate ester transport(GO:1901374) |
| 2.7 | 8.2 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 2.5 | 7.5 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 2.5 | 20.0 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 2.3 | 9.3 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) cellular response to thyroxine stimulus(GO:0097069) |
| 2.3 | 9.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 2.2 | 13.3 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 1.8 | 5.3 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 1.7 | 5.2 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 1.5 | 16.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.4 | 9.8 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 1.3 | 9.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 1.3 | 14.0 | GO:0015747 | urate transport(GO:0015747) |
| 1.3 | 3.8 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 1.0 | 6.9 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.9 | 17.9 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.7 | 5.7 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.7 | 3.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.7 | 7.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.7 | 2.0 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.7 | 5.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.7 | 7.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.6 | 7.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.6 | 13.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.6 | 4.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.6 | 1.8 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 0.6 | 2.9 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.6 | 3.4 | GO:0042908 | canalicular bile acid transport(GO:0015722) xenobiotic transport(GO:0042908) |
| 0.5 | 7.8 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.5 | 2.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.5 | 56.1 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.5 | 6.9 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.5 | 6.8 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.4 | 1.3 | GO:0060221 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) retinal rod cell differentiation(GO:0060221) |
| 0.4 | 1.3 | GO:0032639 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.4 | 1.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.4 | 1.1 | GO:0000958 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) |
| 0.4 | 1.4 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 0.3 | 1.0 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.3 | 1.3 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.3 | 1.6 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.3 | 4.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.3 | 2.4 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.3 | 0.9 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.3 | 1.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 2.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 | 4.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.3 | 13.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.2 | 4.8 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 2.6 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 2.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 9.9 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.2 | 12.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.2 | 2.5 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.2 | 3.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.2 | 1.6 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 4.3 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.1 | 5.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 3.7 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 9.9 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 3.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.4 | GO:0065001 | establishment of anatomical structure orientation(GO:0048560) specification of axis polarity(GO:0065001) |
| 0.1 | 0.5 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 2.9 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.1 | 2.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 3.8 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 2.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 10.3 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 2.8 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 88.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 7.0 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 0.5 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.1 | 1.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.9 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.3 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 2.5 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.5 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.9 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.1 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.0 | 4.3 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 3.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 1.9 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 1.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 1.9 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.7 | 81.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.6 | 11.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 1.5 | 4.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 1.3 | 4.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 1.3 | 3.8 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 1.0 | 5.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.9 | 13.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.8 | 21.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.5 | 27.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.5 | 7.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.4 | 1.3 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.4 | 9.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 6.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 1.1 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.3 | 14.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.3 | 1.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 9.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.3 | 15.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 11.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 1.3 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.2 | 4.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 2.9 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 3.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 10.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 0.9 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.1 | 0.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 2.0 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 2.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 15.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.9 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 26.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 9.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 83.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.9 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 9.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 7.1 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 16.5 | GO:0061695 | transferase complex, transferring phosphorus-containing groups(GO:0061695) |
| 0.1 | 5.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 4.7 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 6.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 21.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 23.1 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 1.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 5.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 50.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 3.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 32.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 2.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 43.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.2 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.4 | 70.1 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 9.8 | 9.8 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 7.6 | 22.9 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 7.6 | 22.7 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 4.2 | 21.1 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 4.2 | 29.5 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 3.8 | 26.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 3.6 | 14.5 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 3.5 | 66.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 3.1 | 21.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.9 | 34.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 2.9 | 20.0 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 2.7 | 8.2 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 2.7 | 48.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 2.6 | 7.8 | GO:0008405 | arachidonic acid 11,12-epoxygenase activity(GO:0008405) |
| 2.6 | 10.3 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 2.3 | 29.6 | GO:0019841 | retinol binding(GO:0019841) |
| 2.3 | 9.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 2.1 | 8.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 2.0 | 16.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.8 | 9.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 1.5 | 4.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.4 | 14.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.4 | 43.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.3 | 4.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 1.0 | 7.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.0 | 5.7 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 1.0 | 3.8 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.8 | 4.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.8 | 10.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.7 | 2.2 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.7 | 9.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.7 | 6.9 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.7 | 17.5 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.7 | 2.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.6 | 4.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.6 | 2.9 | GO:0016918 | retinal binding(GO:0016918) |
| 0.5 | 9.3 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.5 | 13.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 1.3 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.4 | 9.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.4 | 9.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.4 | 10.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 1.9 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.4 | 3.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.4 | 1.1 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.3 | 17.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.3 | 1.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.3 | 9.9 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.3 | 1.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 2.8 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.2 | 1.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.2 | 5.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 3.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 2.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 3.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 2.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 1.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.2 | 2.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 2.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 0.9 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 0.9 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 5.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 1.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 2.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 2.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 2.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 1.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.5 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 7.7 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 21.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 11.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 3.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 30.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 3.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.8 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 52.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 3.2 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 10.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 82.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.7 | 24.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.6 | 14.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.5 | 17.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.4 | 15.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 51.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 3.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 4.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 3.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.9 | 71.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 2.3 | 81.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.9 | 17.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 1.3 | 34.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 1.1 | 21.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.8 | 9.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.6 | 5.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.6 | 15.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 32.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.5 | 12.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.5 | 20.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 18.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.4 | 10.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 21.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.4 | 2.9 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 3.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 3.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 4.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 4.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 5.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 3.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 8.0 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.8 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 5.6 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 1.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 2.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |